Scott W. Linderman
Statistics Department and The Wu Tsai Neurosciences Institute, Stanford University, Stanford, CA, USA
A Bayesian Nonparametric Perspective on Mahalanobis Distance for Out of Distribution Detection
Feb 12, 2025Abstract:Bayesian nonparametric methods are naturally suited to the problem of out-of-distribution (OOD) detection. However, these techniques have largely been eschewed in favor of simpler methods based on distances between pre-trained or learned embeddings of data points. Here we show a formal relationship between Bayesian nonparametric models and the relative Mahalanobis distance score (RMDS), a commonly used method for OOD detection. Building on this connection, we propose Bayesian nonparametric mixture models with hierarchical priors that generalize the RMDS. We evaluate these models on the OpenOOD detection benchmark and show that Bayesian nonparametric methods can improve upon existing OOD methods, especially in regimes where training classes differ in their covariance structure and where there are relatively few data points per class.
Brain-to-Text Benchmark '24: Lessons Learned
Dec 23, 2024

Abstract:Speech brain-computer interfaces aim to decipher what a person is trying to say from neural activity alone, restoring communication to people with paralysis who have lost the ability to speak intelligibly. The Brain-to-Text Benchmark '24 and associated competition was created to foster the advancement of decoding algorithms that convert neural activity to text. Here, we summarize the lessons learned from the competition ending on June 1, 2024 (the top 4 entrants also presented their experiences in a recorded webinar). The largest improvements in accuracy were achieved using an ensembling approach, where the output of multiple independent decoders was merged using a fine-tuned large language model (an approach used by all 3 top entrants). Performance gains were also found by improving how the baseline recurrent neural network (RNN) model was trained, including by optimizing learning rate scheduling and by using a diphone training objective. Improving upon the model architecture itself proved more difficult, however, with attempts to use deep state space models or transformers not yet appearing to offer a benefit over the RNN baseline. The benchmark will remain open indefinitely to support further work towards increasing the accuracy of brain-to-text algorithms.
Towards Scalable and Stable Parallelization of Nonlinear RNNs
Jul 26, 2024



Abstract:Conventional nonlinear RNNs are not naturally parallelizable across the sequence length, whereas transformers and linear RNNs are. Lim et al. [2024] therefore tackle parallelized evaluation of nonlinear RNNs by posing it as a fixed point problem, solved with Newton's method. By deriving and applying a parallelized form of Newton's method, they achieve huge speedups over sequential evaluation. However, their approach inherits cubic computational complexity and numerical instability. We tackle these weaknesses. To reduce the computational complexity, we apply quasi-Newton approximations and show they converge comparably to full-Newton, use less memory, and are faster. To stabilize Newton's method, we leverage a connection between Newton's method damped with trust regions and Kalman smoothing. This connection allows us to stabilize Newtons method, per the trust region, while using efficient parallelized Kalman algorithms to retain performance. We compare these methods empirically, and highlight the use cases where each algorithm excels.
Towards a theory of learning dynamics in deep state space models
Jul 10, 2024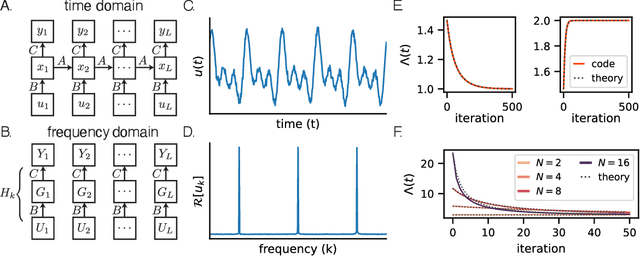
Abstract:State space models (SSMs) have shown remarkable empirical performance on many long sequence modeling tasks, but a theoretical understanding of these models is still lacking. In this work, we study the learning dynamics of linear SSMs to understand how covariance structure in data, latent state size, and initialization affect the evolution of parameters throughout learning with gradient descent. We show that focusing on the learning dynamics in the frequency domain affords analytical solutions under mild assumptions, and we establish a link between one-dimensional SSMs and the dynamics of deep linear feed-forward networks. Finally, we analyze how latent state over-parameterization affects convergence time and describe future work in extending our results to the study of deep SSMs with nonlinear connections. This work is a step toward a theory of learning dynamics in deep state space models.
Convolutional State Space Models for Long-Range Spatiotemporal Modeling
Oct 30, 2023Abstract:Effectively modeling long spatiotemporal sequences is challenging due to the need to model complex spatial correlations and long-range temporal dependencies simultaneously. ConvLSTMs attempt to address this by updating tensor-valued states with recurrent neural networks, but their sequential computation makes them slow to train. In contrast, Transformers can process an entire spatiotemporal sequence, compressed into tokens, in parallel. However, the cost of attention scales quadratically in length, limiting their scalability to longer sequences. Here, we address the challenges of prior methods and introduce convolutional state space models (ConvSSM) that combine the tensor modeling ideas of ConvLSTM with the long sequence modeling approaches of state space methods such as S4 and S5. First, we demonstrate how parallel scans can be applied to convolutional recurrences to achieve subquadratic parallelization and fast autoregressive generation. We then establish an equivalence between the dynamics of ConvSSMs and SSMs, which motivates parameterization and initialization strategies for modeling long-range dependencies. The result is ConvS5, an efficient ConvSSM variant for long-range spatiotemporal modeling. ConvS5 significantly outperforms Transformers and ConvLSTM on a long horizon Moving-MNIST experiment while training 3X faster than ConvLSTM and generating samples 400X faster than Transformers. In addition, ConvS5 matches or exceeds the performance of state-of-the-art methods on challenging DMLab, Minecraft and Habitat prediction benchmarks and enables new directions for modeling long spatiotemporal sequences.
Switching Autoregressive Low-rank Tensor Models
Jun 07, 2023



Abstract:An important problem in time-series analysis is modeling systems with time-varying dynamics. Probabilistic models with joint continuous and discrete latent states offer interpretable, efficient, and experimentally useful descriptions of such data. Commonly used models include autoregressive hidden Markov models (ARHMMs) and switching linear dynamical systems (SLDSs), each with its own advantages and disadvantages. ARHMMs permit exact inference and easy parameter estimation, but are parameter intensive when modeling long dependencies, and hence are prone to overfitting. In contrast, SLDSs can capture long-range dependencies in a parameter efficient way through Markovian latent dynamics, but present an intractable likelihood and a challenging parameter estimation task. In this paper, we propose switching autoregressive low-rank tensor (SALT) models, which retain the advantages of both approaches while ameliorating the weaknesses. SALT parameterizes the tensor of an ARHMM with a low-rank factorization to control the number of parameters and allow longer range dependencies without overfitting. We prove theoretical and discuss practical connections between SALT, linear dynamical systems, and SLDSs. We empirically demonstrate quantitative advantages of SALT models on a range of simulated and real prediction tasks, including behavioral and neural datasets. Furthermore, the learned low-rank tensor provides novel insights into temporal dependencies within each discrete state.
Revisiting Structured Variational Autoencoders
May 25, 2023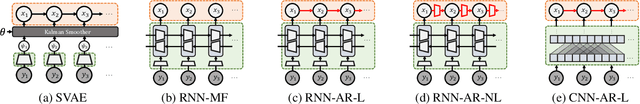
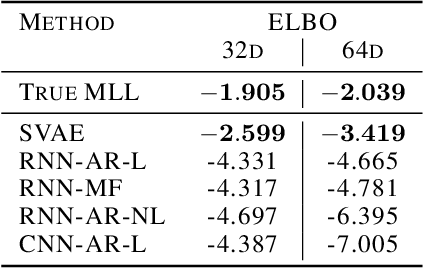
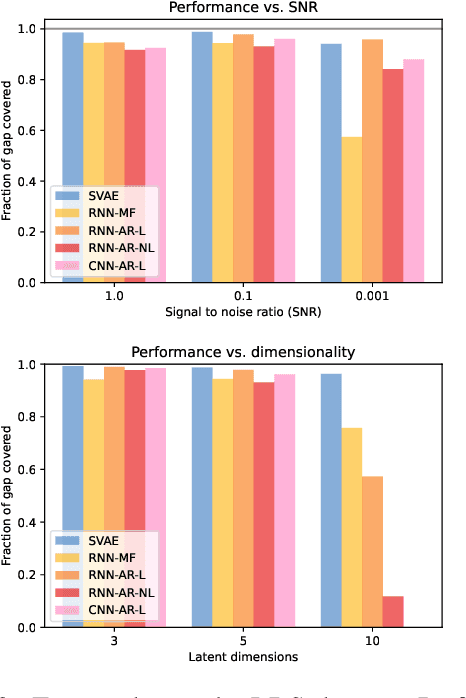
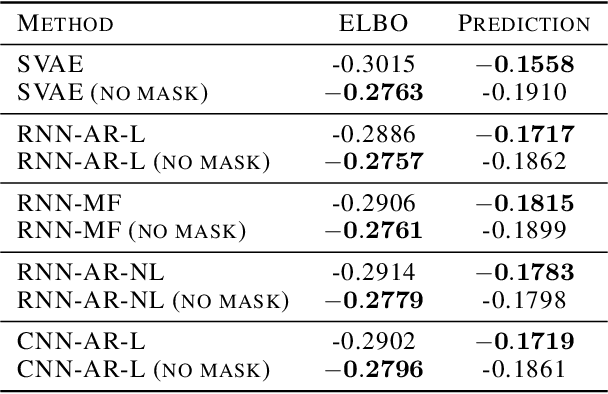
Abstract:Structured variational autoencoders (SVAEs) combine probabilistic graphical model priors on latent variables, deep neural networks to link latent variables to observed data, and structure-exploiting algorithms for approximate posterior inference. These models are particularly appealing for sequential data, where the prior can capture temporal dependencies. However, despite their conceptual elegance, SVAEs have proven difficult to implement, and more general approaches have been favored in practice. Here, we revisit SVAEs using modern machine learning tools and demonstrate their advantages over more general alternatives in terms of both accuracy and efficiency. First, we develop a modern implementation for hardware acceleration, parallelization, and automatic differentiation of the message passing algorithms at the core of the SVAE. Second, we show that by exploiting structure in the prior, the SVAE learns more accurate models and posterior distributions, which translate into improved performance on prediction tasks. Third, we show how the SVAE can naturally handle missing data, and we leverage this ability to develop a novel, self-supervised training approach. Altogether, these results show that the time is ripe to revisit structured variational autoencoders.
Simplified State Space Layers for Sequence Modeling
Aug 09, 2022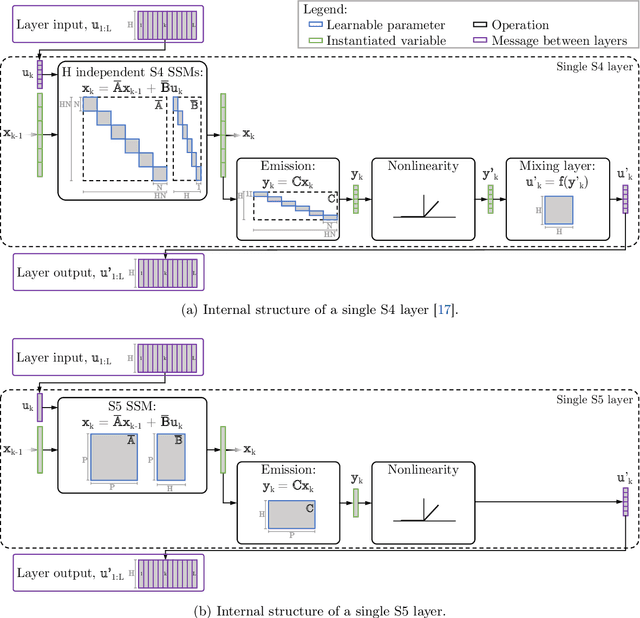
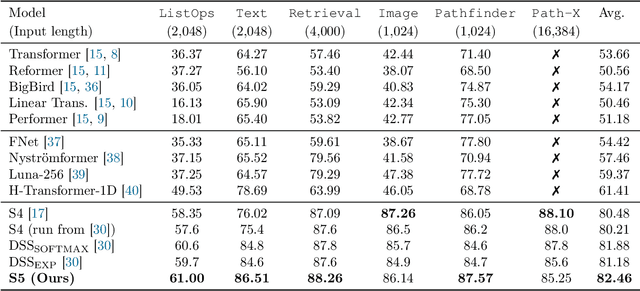
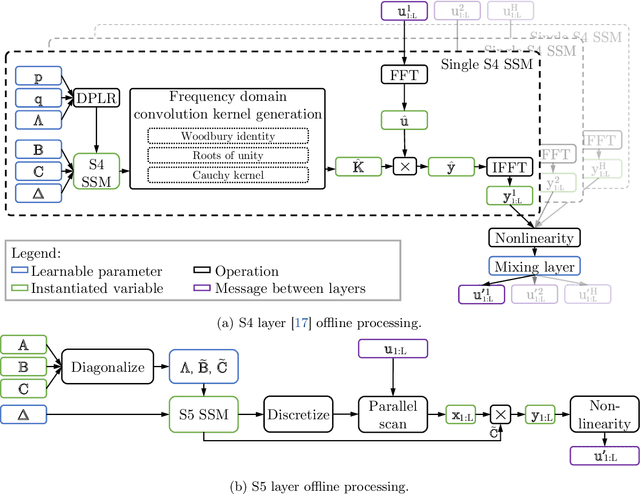
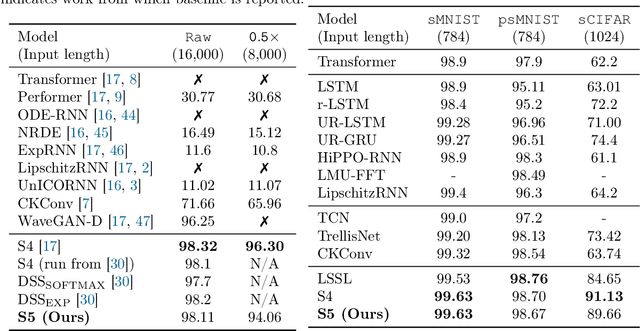
Abstract:Efficiently modeling long-range dependencies is an important goal in sequence modeling. Recently, models using structured state space sequence (S4) layers achieved state-of-the-art performance on many long-range tasks. The S4 layer combines linear state space models (SSMs) with deep learning techniques and leverages the HiPPO framework for online function approximation to achieve high performance. However, this framework led to architectural constraints and computational difficulties that make the S4 approach complicated to understand and implement. We revisit the idea that closely following the HiPPO framework is necessary for high performance. Specifically, we replace the bank of many independent single-input, single-output (SISO) SSMs the S4 layer uses with one multi-input, multi-output (MIMO) SSM with a reduced latent dimension. The reduced latent dimension of the MIMO system allows for the use of efficient parallel scans which simplify the computations required to apply the S5 layer as a sequence-to-sequence transformation. In addition, we initialize the state matrix of the S5 SSM with an approximation to the HiPPO-LegS matrix used by S4's SSMs and show that this serves as an effective initialization for the MIMO setting. S5 matches S4's performance on long-range tasks, including achieving an average of 82.46% on the suite of Long Range Arena benchmarks compared to S4's 80.48% and the best transformer variant's 61.41%.
Spatiotemporal Clustering with Neyman-Scott Processes via Connections to Bayesian Nonparametric Mixture Models
Jan 14, 2022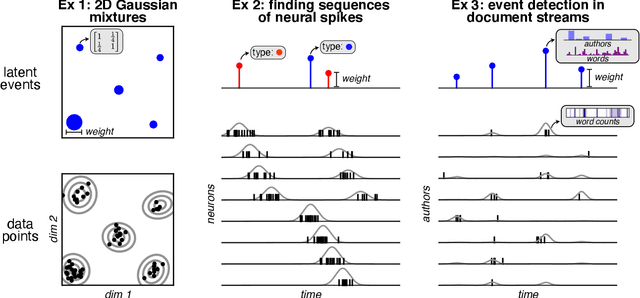
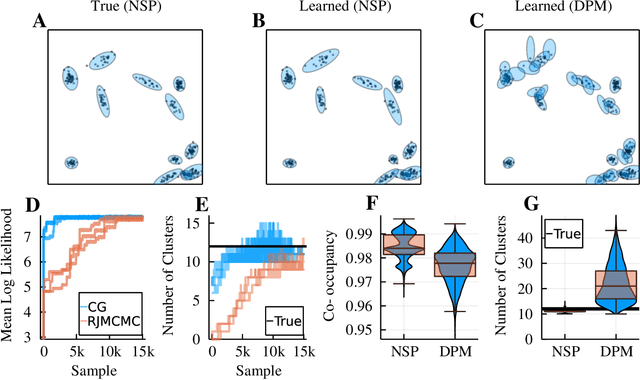
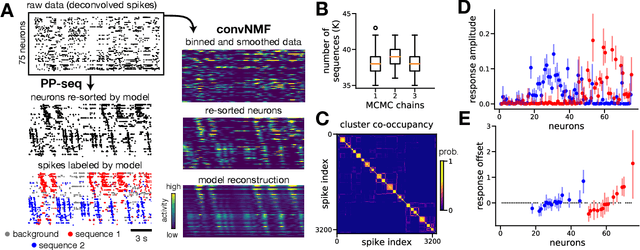
Abstract:Neyman-Scott processes (NSPs) are point process models that generate clusters of points in time or space. They are natural models for a wide range of phenomena, ranging from neural spike trains to document streams. The clustering property is achieved via a doubly stochastic formulation: first, a set of latent events is drawn from a Poisson process; then, each latent event generates a set of observed data points according to another Poisson process. This construction is similar to Bayesian nonparametric mixture models like the Dirichlet process mixture model (DPMM) in that the number of latent events (i.e. clusters) is a random variable, but the point process formulation makes the NSP especially well suited to modeling spatiotemporal data. While many specialized algorithms have been developed for DPMMs, comparatively fewer works have focused on inference in NSPs. Here, we present novel connections between NSPs and DPMMs, with the key link being a third class of Bayesian mixture models called mixture of finite mixture models (MFMMs). Leveraging this connection, we adapt the standard collapsed Gibbs sampling algorithm for DPMMs to enable scalable Bayesian inference on NSP models. We demonstrate the potential of Neyman-Scott processes on a variety of applications including sequence detection in neural spike trains and event detection in document streams.
Reverse engineering recurrent neural networks with Jacobian switching linear dynamical systems
Nov 01, 2021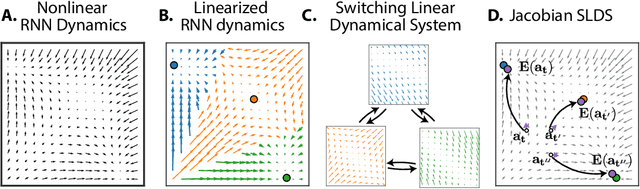
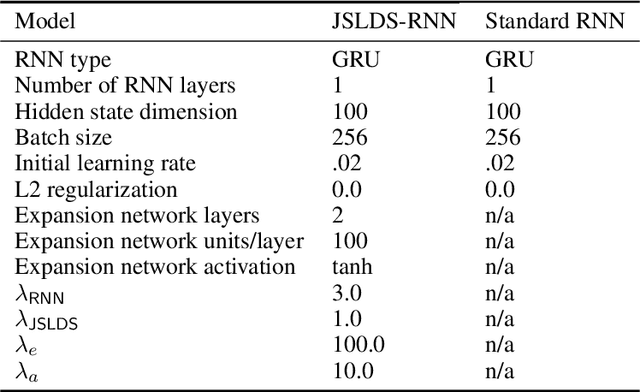
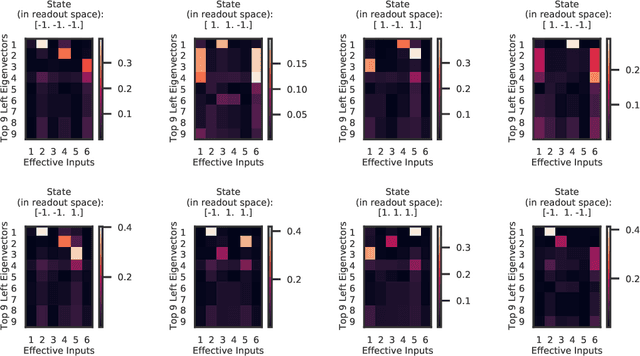
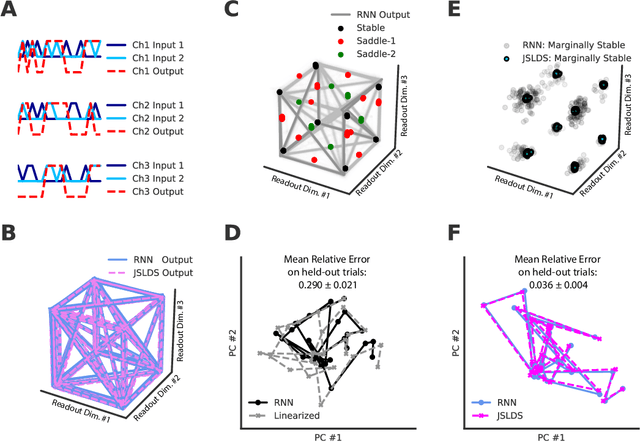
Abstract:Recurrent neural networks (RNNs) are powerful models for processing time-series data, but it remains challenging to understand how they function. Improving this understanding is of substantial interest to both the machine learning and neuroscience communities. The framework of reverse engineering a trained RNN by linearizing around its fixed points has provided insight, but the approach has significant challenges. These include difficulty choosing which fixed point to expand around when studying RNN dynamics and error accumulation when reconstructing the nonlinear dynamics with the linearized dynamics. We present a new model that overcomes these limitations by co-training an RNN with a novel switching linear dynamical system (SLDS) formulation. A first-order Taylor series expansion of the co-trained RNN and an auxiliary function trained to pick out the RNN's fixed points govern the SLDS dynamics. The results are a trained SLDS variant that closely approximates the RNN, an auxiliary function that can produce a fixed point for each point in state-space, and a trained nonlinear RNN whose dynamics have been regularized such that its first-order terms perform the computation, if possible. This model removes the post-training fixed point optimization and allows us to unambiguously study the learned dynamics of the SLDS at any point in state-space. It also generalizes SLDS models to continuous manifolds of switching points while sharing parameters across switches. We validate the utility of the model on two synthetic tasks relevant to previous work reverse engineering RNNs. We then show that our model can be used as a drop-in in more complex architectures, such as LFADS, and apply this LFADS hybrid to analyze single-trial spiking activity from the motor system of a non-human primate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge