Joshua I. Glaser
Switching Autoregressive Low-rank Tensor Models
Jun 07, 2023



Abstract:An important problem in time-series analysis is modeling systems with time-varying dynamics. Probabilistic models with joint continuous and discrete latent states offer interpretable, efficient, and experimentally useful descriptions of such data. Commonly used models include autoregressive hidden Markov models (ARHMMs) and switching linear dynamical systems (SLDSs), each with its own advantages and disadvantages. ARHMMs permit exact inference and easy parameter estimation, but are parameter intensive when modeling long dependencies, and hence are prone to overfitting. In contrast, SLDSs can capture long-range dependencies in a parameter efficient way through Markovian latent dynamics, but present an intractable likelihood and a challenging parameter estimation task. In this paper, we propose switching autoregressive low-rank tensor (SALT) models, which retain the advantages of both approaches while ameliorating the weaknesses. SALT parameterizes the tensor of an ARHMM with a low-rank factorization to control the number of parameters and allow longer range dependencies without overfitting. We prove theoretical and discuss practical connections between SALT, linear dynamical systems, and SLDSs. We empirically demonstrate quantitative advantages of SALT models on a range of simulated and real prediction tasks, including behavioral and neural datasets. Furthermore, the learned low-rank tensor provides novel insights into temporal dependencies within each discrete state.
Deep learning approaches for neural decoding: from CNNs to LSTMs and spikes to fMRI
May 19, 2020


Abstract:Decoding behavior, perception, or cognitive state directly from neural signals has applications in brain-computer interface research as well as implications for systems neuroscience. In the last decade, deep learning has become the state-of-the-art method in many machine learning tasks ranging from speech recognition to image segmentation. The success of deep networks in other domains has led to a new wave of applications in neuroscience. In this article, we review deep learning approaches to neural decoding. We describe the architectures used for extracting useful features from neural recording modalities ranging from spikes to EEG. Furthermore, we explore how deep learning has been leveraged to predict common outputs including movement, speech, and vision, with a focus on how pretrained deep networks can be incorporated as priors for complex decoding targets like acoustic speech or images. Deep learning has been shown to be a useful tool for improving the accuracy and flexibility of neural decoding across a wide range of tasks, and we point out areas for future scientific development.
The Roles of Supervised Machine Learning in Systems Neuroscience
May 21, 2018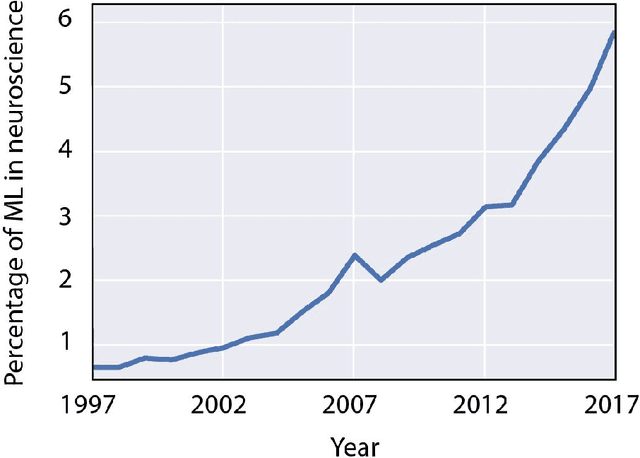
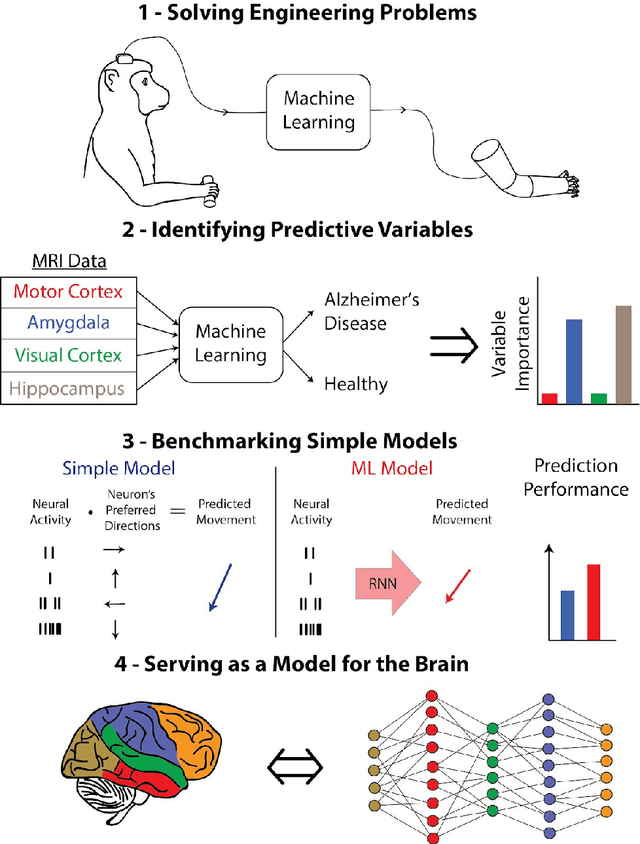
Abstract:Over the last several years, the use of machine learning (ML) in neuroscience has been increasing exponentially. Here, we review ML's contributions, both realized and potential, across several areas of systems neuroscience. We describe four primary roles of ML within neuroscience: 1) creating solutions to engineering problems, 2) identifying predictive variables, 3) setting benchmarks for simple models of the brain, and 4) serving itself as a model for the brain. The breadth and ease of its applicability suggests that machine learning should be in the toolbox of most systems neuroscientists.
Machine learning for neural decoding
May 04, 2018



Abstract:Despite rapid advances in machine learning tools, the majority of neural decoding approaches still use traditional methods. Improving the performance of neural decoding algorithms allows us to better understand the information contained in a neural population, and can help advance engineering applications such as brain machine interfaces. Here, we apply modern machine learning techniques, including neural networks and gradient boosting, to decode from spiking activity in 1) motor cortex, 2) somatosensory cortex, and 3) hippocampus. We compare the predictive ability of these modern methods with traditional decoding methods such as Wiener and Kalman filters. Modern methods, in particular neural networks and ensembles, significantly outperformed the traditional approaches. For instance, for all of the three brain areas, an LSTM decoder explained over 40% of the unexplained variance from a Wiener filter. These results suggest that modern machine learning techniques should become the standard methodology for neural decoding. We provide a tutorial and code to facilitate wider implementation of these methods.
Puzzle Imaging: Using Large-scale Dimensionality Reduction Algorithms for Localization
Jun 21, 2015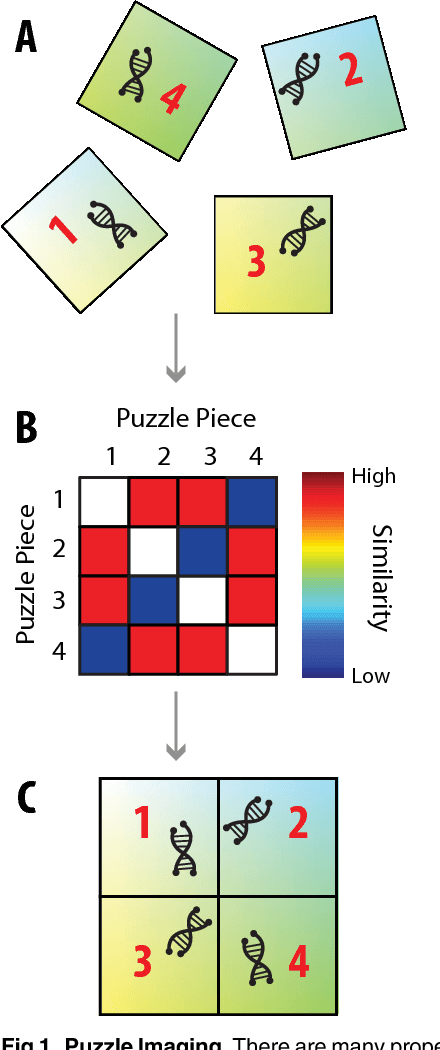

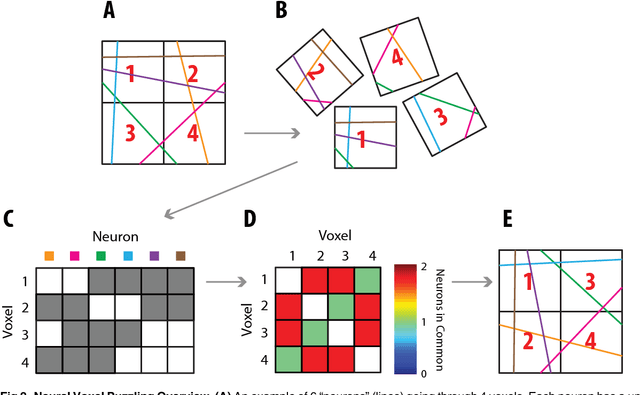

Abstract:Current high-resolution imaging techniques require an intact sample that preserves spatial relationships. We here present a novel approach, "puzzle imaging," that allows imaging a spatially scrambled sample. This technique takes many spatially disordered samples, and then pieces them back together using local properties embedded within the sample. We show that puzzle imaging can efficiently produce high-resolution images using dimensionality reduction algorithms. We demonstrate the theoretical capabilities of puzzle imaging in three biological scenarios, showing that (1) relatively precise 3-dimensional brain imaging is possible; (2) the physical structure of a neural network can often be recovered based only on the neural connectivity matrix; and (3) a chemical map could be reproduced using bacteria with chemosensitive DNA and conjugative transfer. The ability to reconstruct scrambled images promises to enable imaging based on DNA sequencing of homogenized tissue samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge