Lee E. Miller
Capturing cross-session neural population variability through self-supervised identification of consistent neuron ensembles
May 19, 2022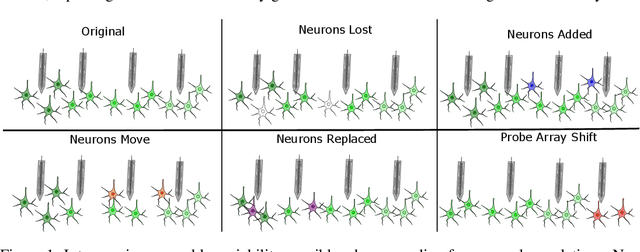
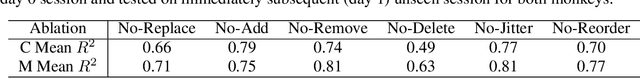

Abstract:Decoding stimuli or behaviour from recorded neural activity is a common approach to interrogate brain function in research, and an essential part of brain-computer and brain-machine interfaces. Reliable decoding even from small neural populations is possible because high dimensional neural population activity typically occupies low dimensional manifolds that are discoverable with suitable latent variable models. Over time however, drifts in activity of individual neurons and instabilities in neural recording devices can be substantial, making stable decoding over days and weeks impractical. While this drift cannot be predicted on an individual neuron level, population level variations over consecutive recording sessions such as differing sets of neurons and varying permutations of consistent neurons in recorded data may be learnable when the underlying manifold is stable over time. Classification of consistent versus unfamiliar neurons across sessions and accounting for deviations in the order of consistent recording neurons in recording datasets over sessions of recordings may then maintain decoding performance. In this work we show that self-supervised training of a deep neural network can be used to compensate for this inter-session variability. As a result, a sequential autoencoding model can maintain state-of-the-art behaviour decoding performance for completely unseen recording sessions several days into the future. Our approach only requires a single recording session for training the model, and is a step towards reliable, recalibration-free brain computer interfaces.
Neural Latents Benchmark '21: Evaluating latent variable models of neural population activity
Sep 10, 2021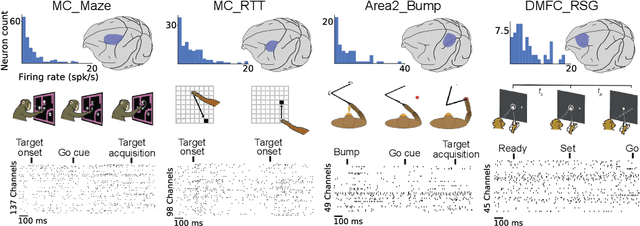
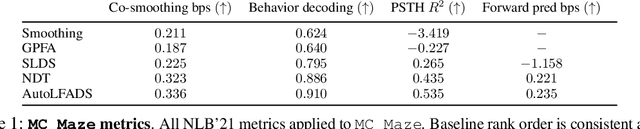
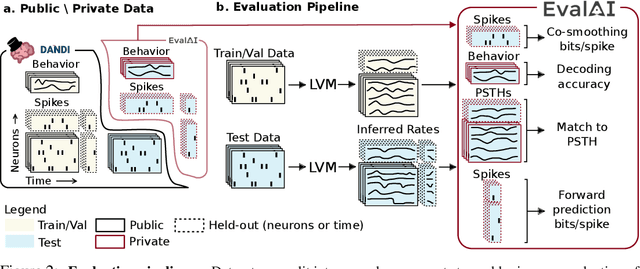
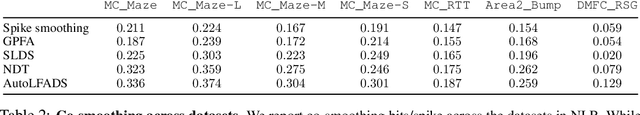
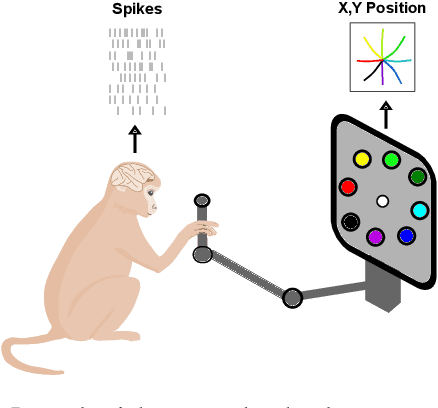
Abstract:Advances in neural recording present increasing opportunities to study neural activity in unprecedented detail. Latent variable models (LVMs) are promising tools for analyzing this rich activity across diverse neural systems and behaviors, as LVMs do not depend on known relationships between the activity and external experimental variables. However, progress in latent variable modeling is currently impeded by a lack of standardization, resulting in methods being developed and compared in an ad hoc manner. To coordinate these modeling efforts, we introduce a benchmark suite for latent variable modeling of neural population activity. We curate four datasets of neural spiking activity from cognitive, sensory, and motor areas to promote models that apply to the wide variety of activity seen across these areas. We identify unsupervised evaluation as a common framework for evaluating models across datasets, and apply several baselines that demonstrate benchmark diversity. We release this benchmark through EvalAI. http://neurallatents.github.io
Adversarial Domain Adaptation for Stable Brain-Machine Interfaces
Sep 28, 2018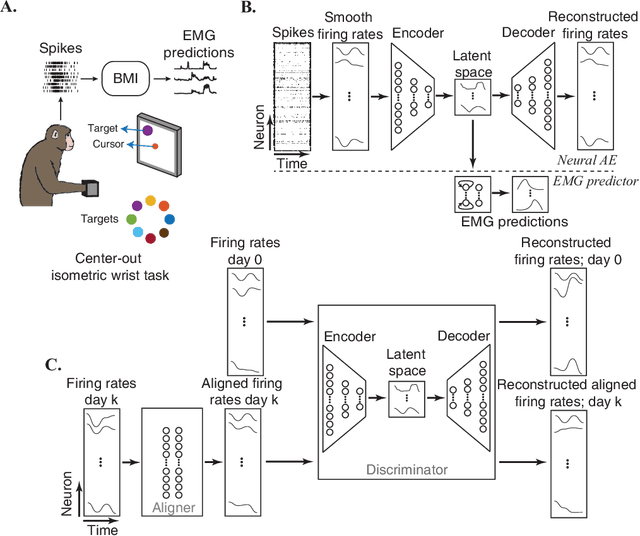
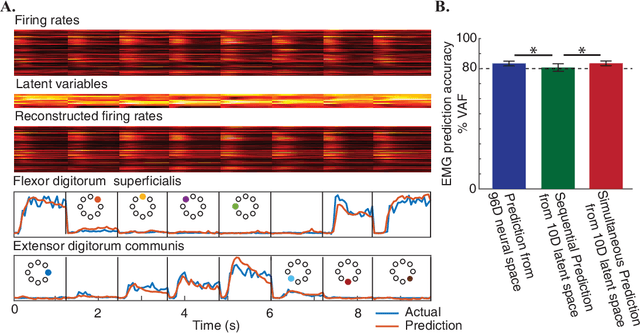
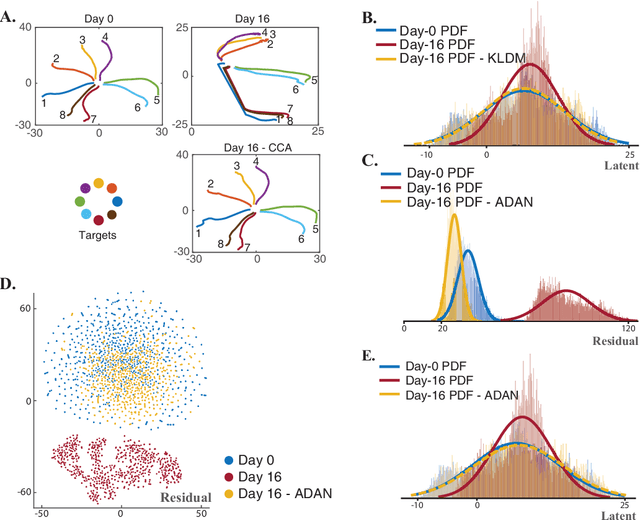
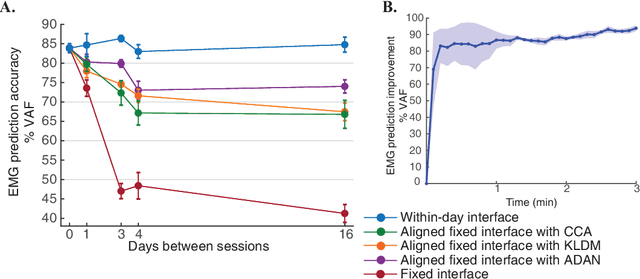
Abstract:Brain-Machine Interfaces (BMIs) have recently emerged as a clinically viable option to restore voluntary movements after paralysis. These devices are based on the ability to extract information about movement intent from neural signals recorded using multi-electrode arrays chronically implanted in the motor cortices of the brain. However, the inherent loss and turnover of recorded neurons requires repeated recalibrations of the interface, which can potentially alter the day-to-day user experience. The resulting need for continued user adaptation interferes with the natural, subconscious use of the BMI. Here, we introduce a new computational approach that decodes movement intent from a low-dimensional latent representation of the neural data. We implement various domain adaptation methods to stabilize the interface over significantly long times. This includes Canonical Correlation Analysis used to align the latent variables across days; this method requires prior point-to-point correspondence of the time series across domains. Alternatively, we match the empirical probability distributions of the latent variables across days through the minimization of their Kullback-Leibler divergence. These two methods provide a significant and comparable improvement in the performance of the interface. However, implementation of an Adversarial Domain Adaptation Network trained to match the empirical probability distribution of the residuals of the reconstructed neural signals outperforms the two methods based on latent variables, while requiring remarkably few data points to solve the domain adaptation problem.
Machine learning for neural decoding
May 04, 2018



Abstract:Despite rapid advances in machine learning tools, the majority of neural decoding approaches still use traditional methods. Improving the performance of neural decoding algorithms allows us to better understand the information contained in a neural population, and can help advance engineering applications such as brain machine interfaces. Here, we apply modern machine learning techniques, including neural networks and gradient boosting, to decode from spiking activity in 1) motor cortex, 2) somatosensory cortex, and 3) hippocampus. We compare the predictive ability of these modern methods with traditional decoding methods such as Wiener and Kalman filters. Modern methods, in particular neural networks and ensembles, significantly outperformed the traditional approaches. For instance, for all of the three brain areas, an LSTM decoder explained over 40% of the unexplained variance from a Wiener filter. These results suggest that modern machine learning techniques should become the standard methodology for neural decoding. We provide a tutorial and code to facilitate wider implementation of these methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge