Matthew T. Kaufman
Deep inference of latent dynamics with spatio-temporal super-resolution using selective backpropagation through time
Oct 29, 2021


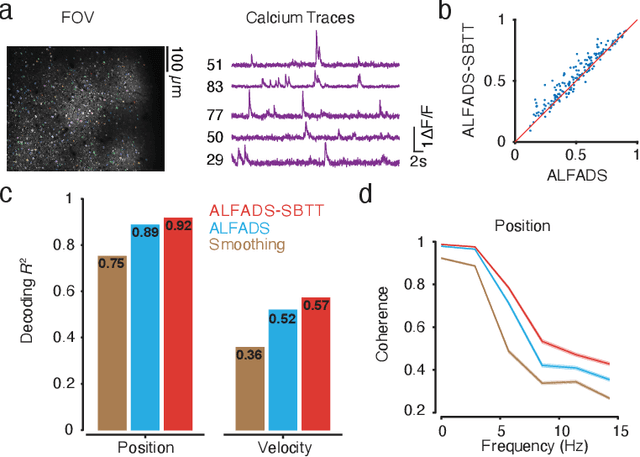
Abstract:Modern neural interfaces allow access to the activity of up to a million neurons within brain circuits. However, bandwidth limits often create a trade-off between greater spatial sampling (more channels or pixels) and the temporal frequency of sampling. Here we demonstrate that it is possible to obtain spatio-temporal super-resolution in neuronal time series by exploiting relationships among neurons, embedded in latent low-dimensional population dynamics. Our novel neural network training strategy, selective backpropagation through time (SBTT), enables learning of deep generative models of latent dynamics from data in which the set of observed variables changes at each time step. The resulting models are able to infer activity for missing samples by combining observations with learned latent dynamics. We test SBTT applied to sequential autoencoders and demonstrate more efficient and higher-fidelity characterization of neural population dynamics in electrophysiological and calcium imaging data. In electrophysiology, SBTT enables accurate inference of neuronal population dynamics with lower interface bandwidths, providing an avenue to significant power savings for implanted neuroelectronic interfaces. In applications to two-photon calcium imaging, SBTT accurately uncovers high-frequency temporal structure underlying neural population activity, substantially outperforming the current state-of-the-art. Finally, we demonstrate that performance could be further improved by using limited, high-bandwidth sampling to pretrain dynamics models, and then using SBTT to adapt these models for sparsely-sampled data.
Neural Latents Benchmark '21: Evaluating latent variable models of neural population activity
Sep 10, 2021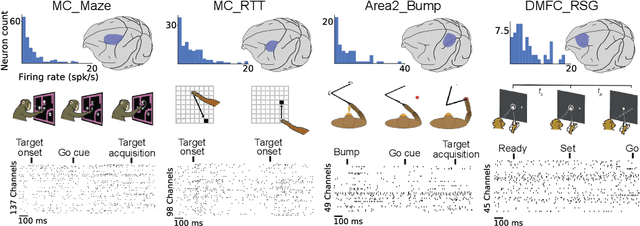
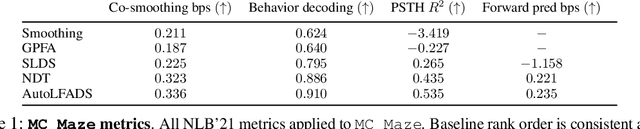
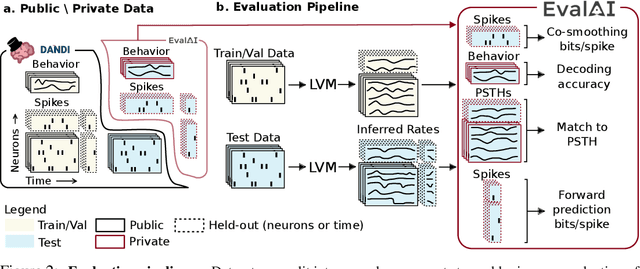
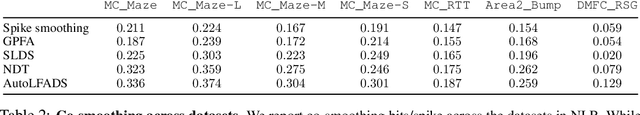
Abstract:Advances in neural recording present increasing opportunities to study neural activity in unprecedented detail. Latent variable models (LVMs) are promising tools for analyzing this rich activity across diverse neural systems and behaviors, as LVMs do not depend on known relationships between the activity and external experimental variables. However, progress in latent variable modeling is currently impeded by a lack of standardization, resulting in methods being developed and compared in an ad hoc manner. To coordinate these modeling efforts, we introduce a benchmark suite for latent variable modeling of neural population activity. We curate four datasets of neural spiking activity from cognitive, sensory, and motor areas to promote models that apply to the wide variety of activity seen across these areas. We identify unsupervised evaluation as a common framework for evaluating models across datasets, and apply several baselines that demonstrate benchmark diversity. We release this benchmark through EvalAI. http://neurallatents.github.io
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge