Nauman Ahad
Relax, it doesn't matter how you get there: A new self-supervised approach for multi-timescale behavior analysis
Mar 15, 2023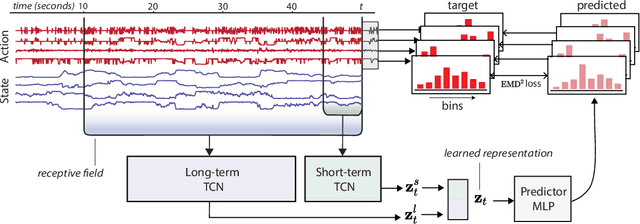
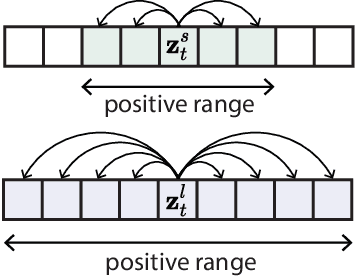
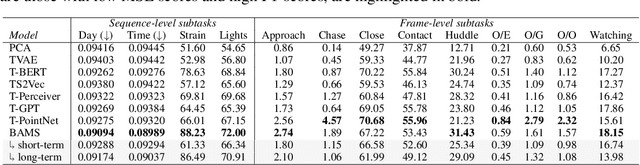
Abstract:Natural behavior consists of dynamics that are complex and unpredictable, especially when trying to predict many steps into the future. While some success has been found in building representations of behavior under constrained or simplified task-based conditions, many of these models cannot be applied to free and naturalistic settings where behavior becomes increasingly hard to model. In this work, we develop a multi-task representation learning model for behavior that combines two novel components: (i) An action prediction objective that aims to predict the distribution of actions over future timesteps, and (ii) A multi-scale architecture that builds separate latent spaces to accommodate short- and long-term dynamics. After demonstrating the ability of the method to build representations of both local and global dynamics in realistic robots in varying environments and terrains, we apply our method to the MABe 2022 Multi-agent behavior challenge, where our model ranks 1st overall and on all global tasks, and 1st or 2nd on 7 out of 9 frame-level tasks. In all of these cases, we show that our model can build representations that capture the many different factors that drive behavior and solve a wide range of downstream tasks.
MTNeuro: A Benchmark for Evaluating Representations of Brain Structure Across Multiple Levels of Abstraction
Jan 01, 2023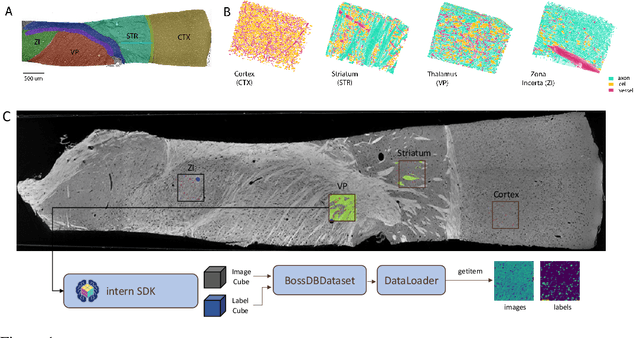
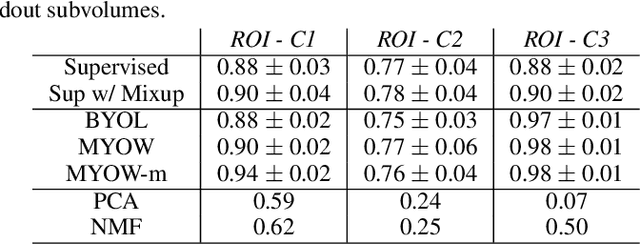
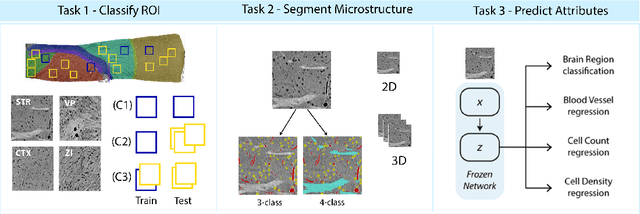
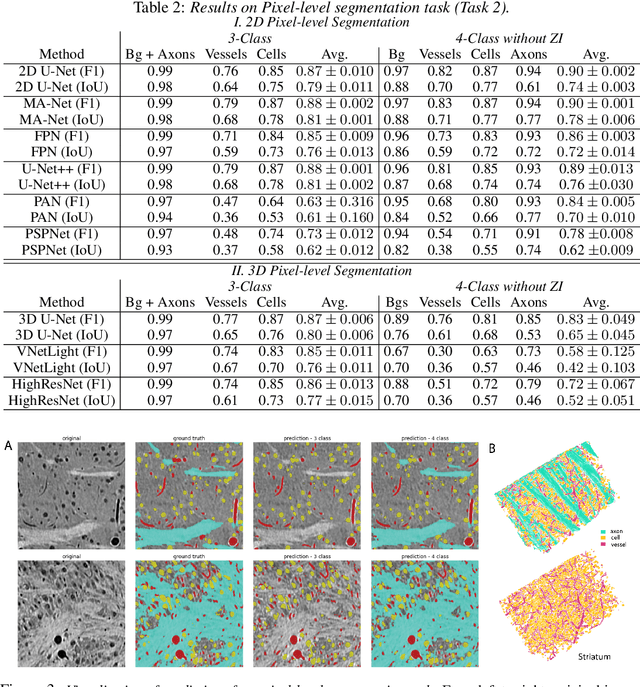
Abstract:There are multiple scales of abstraction from which we can describe the same image, depending on whether we are focusing on fine-grained details or a more global attribute of the image. In brain mapping, learning to automatically parse images to build representations of both small-scale features (e.g., the presence of cells or blood vessels) and global properties of an image (e.g., which brain region the image comes from) is a crucial and open challenge. However, most existing datasets and benchmarks for neuroanatomy consider only a single downstream task at a time. To bridge this gap, we introduce a new dataset, annotations, and multiple downstream tasks that provide diverse ways to readout information about brain structure and architecture from the same image. Our multi-task neuroimaging benchmark (MTNeuro) is built on volumetric, micrometer-resolution X-ray microtomography images spanning a large thalamocortical section of mouse brain, encompassing multiple cortical and subcortical regions. We generated a number of different prediction challenges and evaluated several supervised and self-supervised models for brain-region prediction and pixel-level semantic segmentation of microstructures. Our experiments not only highlight the rich heterogeneity of this dataset, but also provide insights into how self-supervised approaches can be used to learn representations that capture multiple attributes of a single image and perform well on a variety of downstream tasks. Datasets, code, and pre-trained baseline models are provided at: https://mtneuro.github.io/ .
Learning Behavior Representations Through Multi-Timescale Bootstrapping
Jun 14, 2022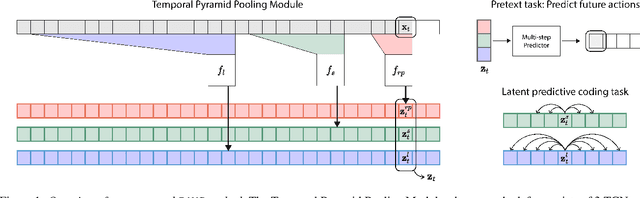
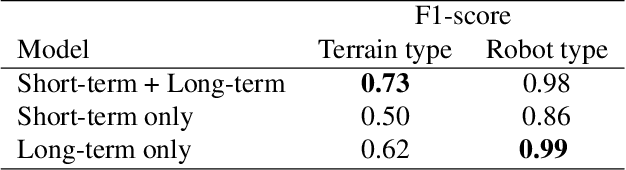
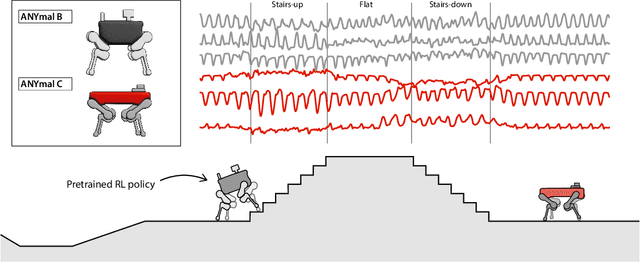
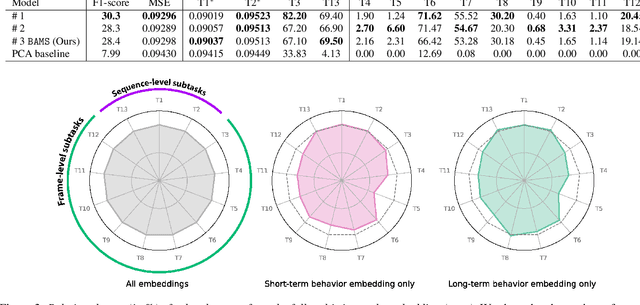
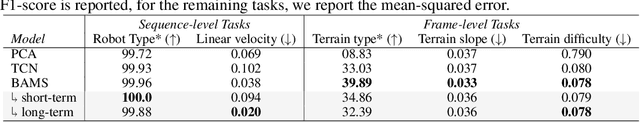
Abstract:Natural behavior consists of dynamics that are both unpredictable, can switch suddenly, and unfold over many different timescales. While some success has been found in building representations of behavior under constrained or simplified task-based conditions, many of these models cannot be applied to free and naturalistic settings due to the fact that they assume a single scale of temporal dynamics. In this work, we introduce Bootstrap Across Multiple Scales (BAMS), a multi-scale representation learning model for behavior: we combine a pooling module that aggregates features extracted over encoders with different temporal receptive fields, and design a set of latent objectives to bootstrap the representations in each respective space to encourage disentanglement across different timescales. We first apply our method on a dataset of quadrupeds navigating in different terrain types, and show that our model captures the temporal complexity of behavior. We then apply our method to the MABe 2022 Multi-agent behavior challenge, where our model ranks 3rd overall and 1st on two subtasks, and show the importance of incorporating multi-timescales when analyzing behavior.
Learning Sinkhorn divergences for supervised change point detection
Feb 10, 2022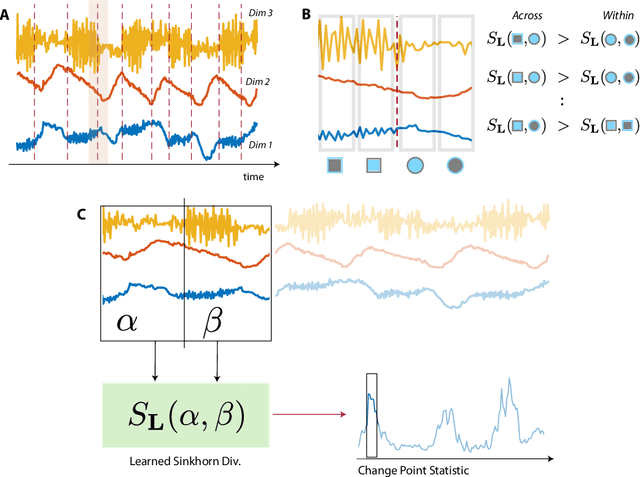
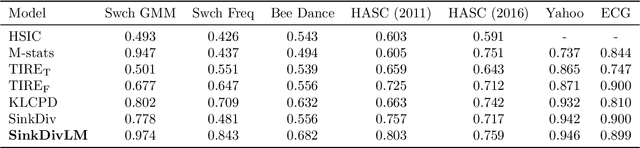
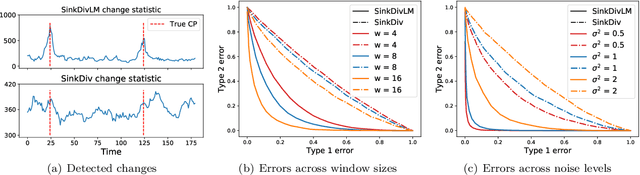
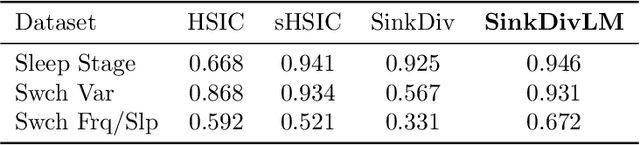
Abstract:Many modern applications require detecting change points in complex sequential data. Most existing methods for change point detection are unsupervised and, as a consequence, lack any information regarding what kind of changes we want to detect or if some kinds of changes are safe to ignore. This often results in poor change detection performance. We present a novel change point detection framework that uses true change point instances as supervision for learning a ground metric such that Sinkhorn divergences can be then used in two-sample tests on sliding windows to detect change points in an online manner. Our method can be used to learn a sparse metric which can be useful for both feature selection and interpretation in high-dimensional change point detection settings. Experiments on simulated as well as real world sequences show that our proposed method can substantially improve change point detection performance over existing unsupervised change point detection methods using only few labeled change point instances.
Deep inference of latent dynamics with spatio-temporal super-resolution using selective backpropagation through time
Oct 29, 2021


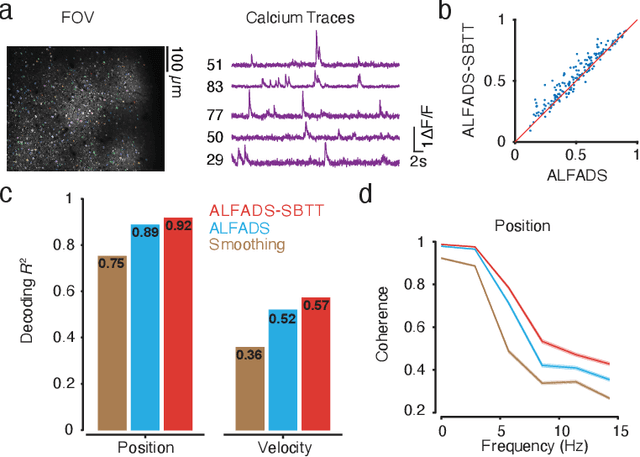
Abstract:Modern neural interfaces allow access to the activity of up to a million neurons within brain circuits. However, bandwidth limits often create a trade-off between greater spatial sampling (more channels or pixels) and the temporal frequency of sampling. Here we demonstrate that it is possible to obtain spatio-temporal super-resolution in neuronal time series by exploiting relationships among neurons, embedded in latent low-dimensional population dynamics. Our novel neural network training strategy, selective backpropagation through time (SBTT), enables learning of deep generative models of latent dynamics from data in which the set of observed variables changes at each time step. The resulting models are able to infer activity for missing samples by combining observations with learned latent dynamics. We test SBTT applied to sequential autoencoders and demonstrate more efficient and higher-fidelity characterization of neural population dynamics in electrophysiological and calcium imaging data. In electrophysiology, SBTT enables accurate inference of neuronal population dynamics with lower interface bandwidths, providing an avenue to significant power savings for implanted neuroelectronic interfaces. In applications to two-photon calcium imaging, SBTT accurately uncovers high-frequency temporal structure underlying neural population activity, substantially outperforming the current state-of-the-art. Finally, we demonstrate that performance could be further improved by using limited, high-bandwidth sampling to pretrain dynamics models, and then using SBTT to adapt these models for sparsely-sampled data.
Semi-supervised sequence classification through change point detection
Oct 06, 2020



Abstract:Sequential sensor data is generated in a wide variety of practical applications. A fundamental challenge involves learning effective classifiers for such sequential data. While deep learning has led to impressive performance gains in recent years in domains such as speech, this has relied on the availability of large datasets of sequences with high-quality labels. In many applications, however, the associated class labels are often extremely limited, with precise labelling/segmentation being too expensive to perform at a high volume. However, large amounts of unlabeled data may still be available. In this paper we propose a novel framework for semi-supervised learning in such contexts. In an unsupervised manner, change point detection methods can be used to identify points within a sequence corresponding to likely class changes. We show that change points provide examples of similar/dissimilar pairs of sequences which, when coupled with labeled, can be used in a semi-supervised classification setting. Leveraging the change points and labeled data, we form examples of similar/dissimilar sequences to train a neural network to learn improved representations for classification. We provide extensive synthetic simulations and show that the learned representations are superior to those learned through an autoencoder and obtain improved results on both simulated and real-world human activity recognition datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge