Alex H. Williams
Quasi Monte Carlo methods enable extremely low-dimensional deep generative models
Jan 26, 2026Abstract:This paper introduces quasi-Monte Carlo latent variable models (QLVMs): a class of deep generative models that are specialized for finding extremely low-dimensional and interpretable embeddings of high-dimensional datasets. Unlike standard approaches, which rely on a learned encoder and variational lower bounds, QLVMs directly approximate the marginal likelihood by randomized quasi-Monte Carlo integration. While this brute force approach has drawbacks in higher-dimensional spaces, we find that it excels in fitting one, two, and three dimensional deep latent variable models. Empirical results on a range of datasets show that QLVMs consistently outperform conventional variational autoencoders (VAEs) and importance weighted autoencoders (IWAEs) with matched latent dimensionality. The resulting embeddings enable transparent visualization and post hoc analyses such as nonparametric density estimation, clustering, and geodesic path computation, which are nontrivial to validate in higher-dimensional spaces. While our approach is compute-intensive and struggles to generate fine-scale details in complex datasets, it offers a compelling solution for applications prioritizing interpretability and latent space analysis.
Modeling Neural Activity with Conditionally Linear Dynamical Systems
Feb 25, 2025



Abstract:Neural population activity exhibits complex, nonlinear dynamics, varying in time, over trials, and across experimental conditions. Here, we develop Conditionally Linear Dynamical System (CLDS) models as a general-purpose method to characterize these dynamics. These models use Gaussian Process (GP) priors to capture the nonlinear dependence of circuit dynamics on task and behavioral variables. Conditioned on these covariates, the data is modeled with linear dynamics. This allows for transparent interpretation and tractable Bayesian inference. We find that CLDS models can perform well even in severely data-limited regimes (e.g. one trial per condition) due to their Bayesian formulation and ability to share statistical power across nearby task conditions. In example applications, we apply CLDS to model thalamic neurons that nonlinearly encode heading direction and to model motor cortical neurons during a cued reaching task
Comparing noisy neural population dynamics using optimal transport distances
Dec 19, 2024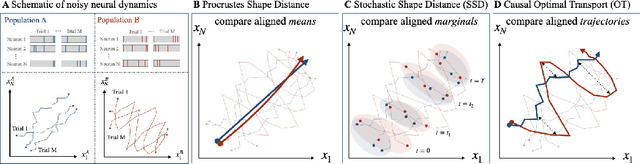
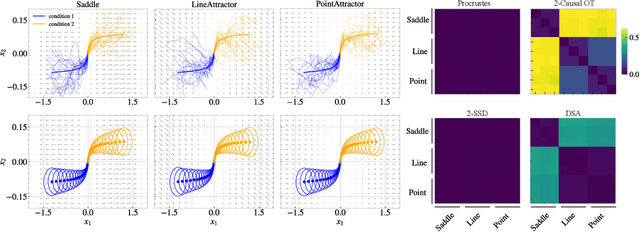
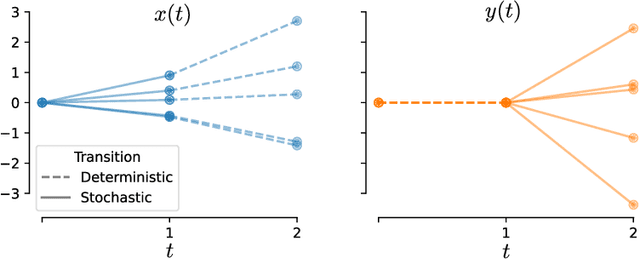
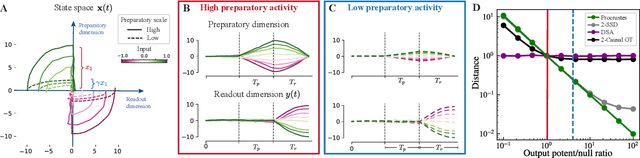
Abstract:Biological and artificial neural systems form high-dimensional neural representations that underpin their computational capabilities. Methods for quantifying geometric similarity in neural representations have become a popular tool for identifying computational principles that are potentially shared across neural systems. These methods generally assume that neural responses are deterministic and static. However, responses of biological systems, and some artificial systems, are noisy and dynamically unfold over time. Furthermore, these characteristics can have substantial influence on a system's computational capabilities. Here, we demonstrate that existing metrics can fail to capture key differences between neural systems with noisy dynamic responses. We then propose a metric for comparing the geometry of noisy neural trajectories, which can be derived as an optimal transport distance between Gaussian processes. We use the metric to compare models of neural responses in different regions of the motor system and to compare the dynamics of latent diffusion models for text-to-image synthesis.
What Representational Similarity Measures Imply about Decodable Information
Nov 12, 2024Abstract:Neural responses encode information that is useful for a variety of downstream tasks. A common approach to understand these systems is to build regression models or ``decoders'' that reconstruct features of the stimulus from neural responses. Popular neural network similarity measures like centered kernel alignment (CKA), canonical correlation analysis (CCA), and Procrustes shape distance, do not explicitly leverage this perspective and instead highlight geometric invariances to orthogonal or affine transformations when comparing representations. Here, we show that many of these measures can, in fact, be equivalently motivated from a decoding perspective. Specifically, measures like CKA and CCA quantify the average alignment between optimal linear readouts across a distribution of decoding tasks. We also show that the Procrustes shape distance upper bounds the distance between optimal linear readouts and that the converse holds for representations with low participation ratio. Overall, our work demonstrates a tight link between the geometry of neural representations and the ability to linearly decode information. This perspective suggests new ways of measuring similarity between neural systems and also provides novel, unifying interpretations of existing measures.
Discriminating image representations with principal distortions
Oct 20, 2024



Abstract:Image representations (artificial or biological) are often compared in terms of their global geometry; however, representations with similar global structure can have strikingly different local geometries. Here, we propose a framework for comparing a set of image representations in terms of their local geometries. We quantify the local geometry of a representation using the Fisher information matrix, a standard statistical tool for characterizing the sensitivity to local stimulus distortions, and use this as a substrate for a metric on the local geometry in the vicinity of a base image. This metric may then be used to optimally differentiate a set of models, by finding a pair of "principal distortions" that maximize the variance of the models under this metric. We use this framework to compare a set of simple models of the early visual system, identifying a novel set of image distortions that allow immediate comparison of the models by visual inspection. In a second example, we apply our method to a set of deep neural network models and reveal differences in the local geometry that arise due to architecture and training types. These examples highlight how our framework can be used to probe for informative differences in local sensitivities between complex computational models, and suggest how it could be used to compare model representations with human perception.
Tensor Decomposition Meets RKHS: Efficient Algorithms for Smooth and Misaligned Data
Aug 11, 2024Abstract:The canonical polyadic (CP) tensor decomposition decomposes a multidimensional data array into a sum of outer products of finite-dimensional vectors. Instead, we can replace some or all of the vectors with continuous functions (infinite-dimensional vectors) from a reproducing kernel Hilbert space (RKHS). We refer to tensors with some infinite-dimensional modes as quasitensors, and the approach of decomposing a tensor with some continuous RKHS modes is referred to as CP-HiFi (hybrid infinite and finite dimensional) tensor decomposition. An advantage of CP-HiFi is that it can enforce smoothness in the infinite dimensional modes. Further, CP-HiFi does not require the observed data to lie on a regular and finite rectangular grid and naturally incorporates misaligned data. We detail the methodology and illustrate it on a synthetic example.
Duality of Bures and Shape Distances with Implications for Comparing Neural Representations
Nov 19, 2023Abstract:A multitude of (dis)similarity measures between neural network representations have been proposed, resulting in a fragmented research landscape. Most of these measures fall into one of two categories. First, measures such as linear regression, canonical correlations analysis (CCA), and shape distances, all learn explicit mappings between neural units to quantify similarity while accounting for expected invariances. Second, measures such as representational similarity analysis (RSA), centered kernel alignment (CKA), and normalized Bures similarity (NBS) all quantify similarity in summary statistics, such as stimulus-by-stimulus kernel matrices, which are already invariant to expected symmetries. Here, we take steps towards unifying these two broad categories of methods by observing that the cosine of the Riemannian shape distance (from category 1) is equal to NBS (from category 2). We explore how this connection leads to new interpretations of shape distances and NBS, and draw contrasts of these measures with CKA, a popular similarity measure in the deep learning literature.
Soft Matching Distance: A metric on neural representations that captures single-neuron tuning
Nov 16, 2023Abstract:Common measures of neural representational (dis)similarity are designed to be insensitive to rotations and reflections of the neural activation space. Motivated by the premise that the tuning of individual units may be important, there has been recent interest in developing stricter notions of representational (dis)similarity that require neurons to be individually matched across networks. When two networks have the same size (i.e. same number of neurons), a distance metric can be formulated by optimizing over neuron index permutations to maximize tuning curve alignment. However, it is not clear how to generalize this metric to measure distances between networks with different sizes. Here, we leverage a connection to optimal transport theory to derive a natural generalization based on "soft" permutations. The resulting metric is symmetric, satisfies the triangle inequality, and can be interpreted as a Wasserstein distance between two empirical distributions. Further, our proposed metric avoids counter-intuitive outcomes suffered by alternative approaches, and captures complementary geometric insights into neural representations that are entirely missed by rotation-invariant metrics.
Estimating Shape Distances on Neural Representations with Limited Samples
Oct 09, 2023Abstract:Measuring geometric similarity between high-dimensional network representations is a topic of longstanding interest to neuroscience and deep learning. Although many methods have been proposed, only a few works have rigorously analyzed their statistical efficiency or quantified estimator uncertainty in data-limited regimes. Here, we derive upper and lower bounds on the worst-case convergence of standard estimators of shape distance$\unicode{x2014}$a measure of representational dissimilarity proposed by Williams et al. (2021). These bounds reveal the challenging nature of the problem in high-dimensional feature spaces. To overcome these challenges, we introduce a new method-of-moments estimator with a tunable bias-variance tradeoff. We show that this estimator achieves superior performance to standard estimators in simulation and on neural data, particularly in high-dimensional settings. Thus, we lay the foundation for a rigorous statistical theory for high-dimensional shape analysis, and we contribute a new estimation method that is well-suited to practical scientific settings.
Representational dissimilarity metric spaces for stochastic neural networks
Nov 21, 2022Abstract:Quantifying similarity between neural representations -- e.g. hidden layer activation vectors -- is a perennial problem in deep learning and neuroscience research. Existing methods compare deterministic responses (e.g. artificial networks that lack stochastic layers) or averaged responses (e.g., trial-averaged firing rates in biological data). However, these measures of deterministic representational similarity ignore the scale and geometric structure of noise, both of which play important roles in neural computation. To rectify this, we generalize previously proposed shape metrics (Williams et al. 2021) to quantify differences in stochastic representations. These new distances satisfy the triangle inequality, and thus can be used as a rigorous basis for many supervised and unsupervised analyses. Leveraging this novel framework, we find that the stochastic geometries of neurobiological representations of oriented visual gratings and naturalistic scenes respectively resemble untrained and trained deep network representations. Further, we are able to more accurately predict certain network attributes (e.g. training hyperparameters) from its position in stochastic (versus deterministic) shape space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge