Michael Aertsen
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022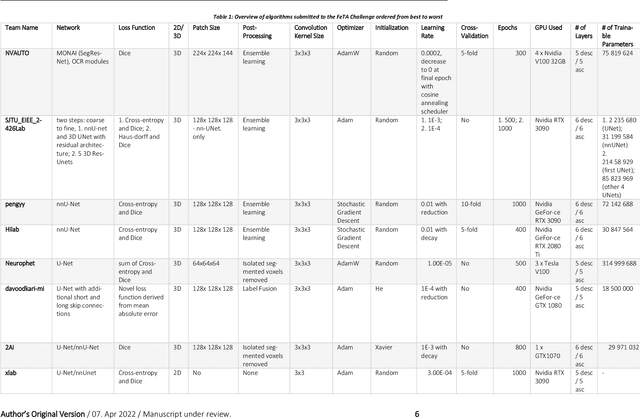
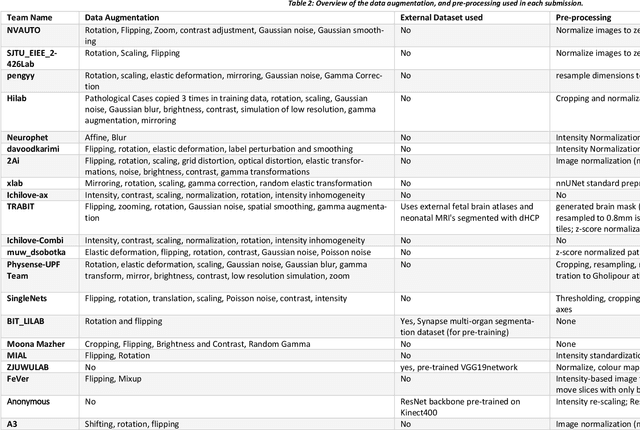
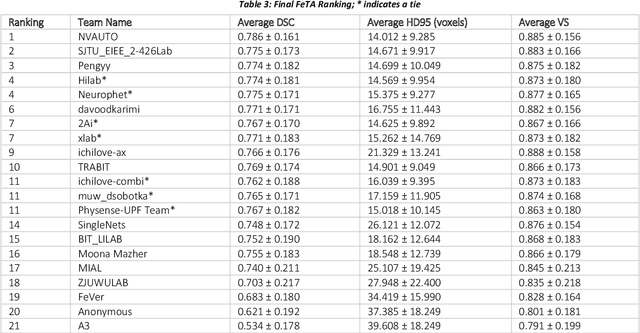
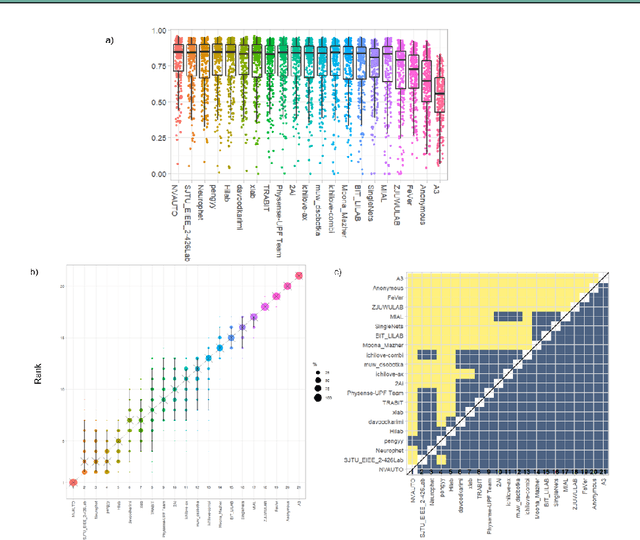
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
A Dempster-Shafer approach to trustworthy AI with application to fetal brain MRI segmentation
Apr 05, 2022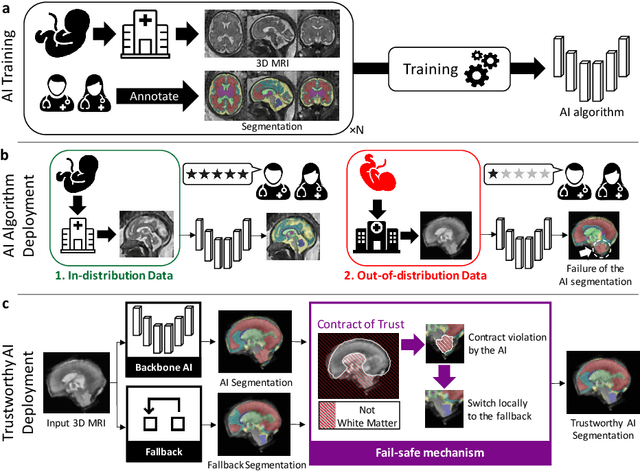
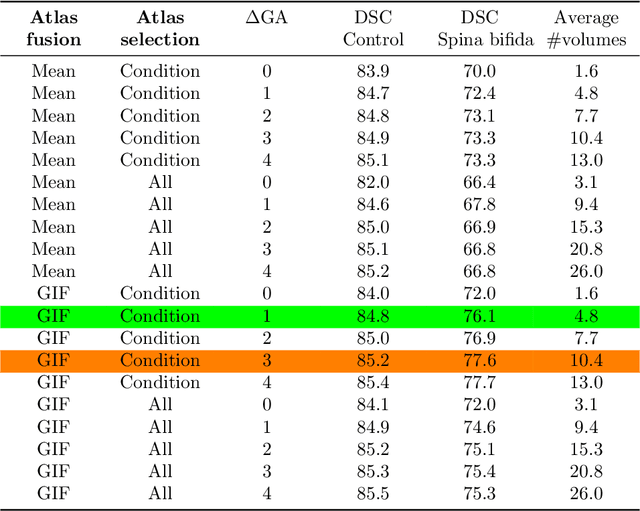
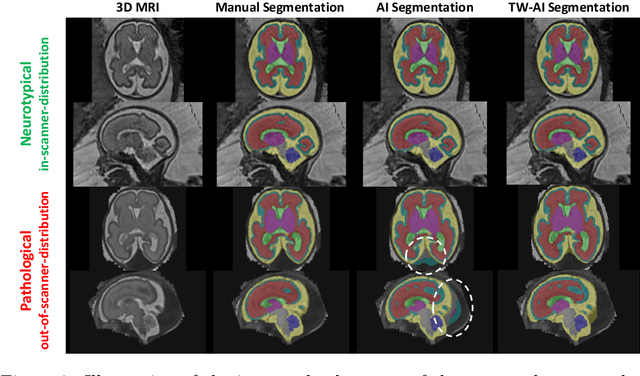
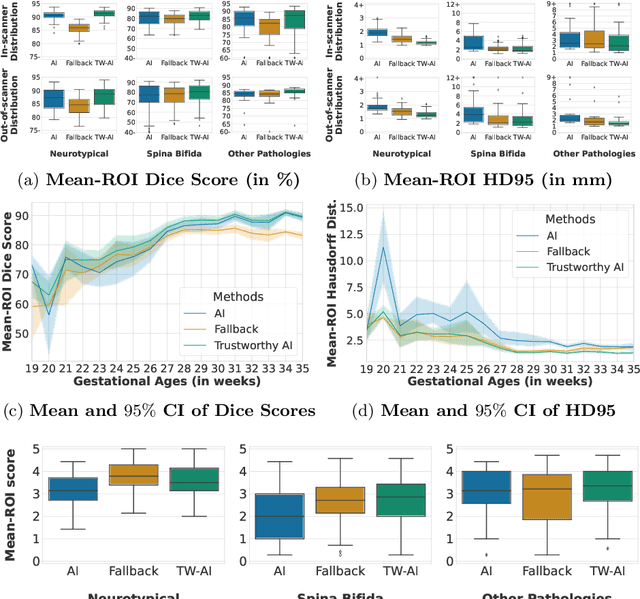
Abstract:Deep learning models for medical image segmentation can fail unexpectedly and spectacularly for pathological cases and for images acquired at different centers than those used for training, with labeling errors that violate expert knowledge about the anatomy and the intensity distribution of the regions to be segmented. Such errors undermine the trustworthiness of deep learning models developed for medical image segmentation. Mechanisms with a fallback method for detecting and correcting such failures are essential for safely translating this technology into clinics and are likely to be a requirement of future regulations on artificial intelligence (AI). Here, we propose a principled trustworthy AI theoretical framework and a practical system that can augment any backbone AI system using a fallback method and a fail-safe mechanism based on Dempster-Shafer theory. Our approach relies on an actionable definition of trustworthy AI. Our method automatically discards the voxel-level labeling predicted by the backbone AI that are likely to violate expert knowledge and relies on a fallback atlas-based segmentation method for those voxels. We demonstrate the effectiveness of the proposed trustworthy AI approach on the largest reported annotated dataset of fetal T2w MRI consisting of 540 manually annotated fetal brain 3D MRIs with neurotypical or abnormal brain development and acquired from 13 sources of data across 6 countries. We show that our trustworthy AI method improves the robustness of a state-of-the-art backbone AI for fetal brain MRI segmentation on MRIs acquired across various centers and for fetuses with various brain abnormalities.
Partial supervision for the FeTA challenge 2021
Nov 03, 2021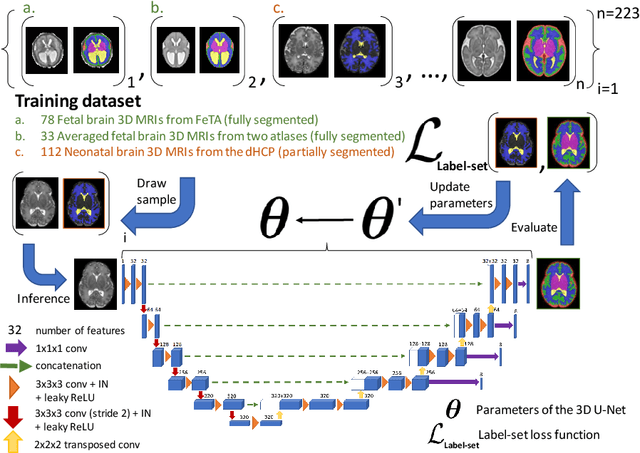
Abstract:This paper describes our method for our participation in the FeTA challenge2021 (team name: TRABIT). The performance of convolutional neural networks for medical image segmentation is thought to correlate positively with the number of training data. The FeTA challenge does not restrict participants to using only the provided training data but also allows for using other publicly available sources. Yet, open access fetal brain data remains limited. An advantageous strategy could thus be to expand the training data to cover broader perinatal brain imaging sources. Perinatal brain MRIs, other than the FeTA challenge data, that are currently publicly available, span normal and pathological fetal atlases as well as neonatal scans. However, perinatal brain MRIs segmented in different datasets typically come with different annotation protocols. This makes it challenging to combine those datasets to train a deep neural network. We recently proposed a family of loss functions, the label-set loss functions, for partially supervised learning. Label-set loss functions allow to train deep neural networks with partially segmented images, i.e. segmentations in which some classes may be grouped into super-classes. We propose to use label-set loss functions to improve the segmentation performance of a state-of-the-art deep learning pipeline for multi-class fetal brain segmentation by merging several publicly available datasets. To promote generalisability, our approach does not introduce any additional hyper-parameters tuning.
Distributionally Robust Segmentation of Abnormal Fetal Brain 3D MRI
Aug 09, 2021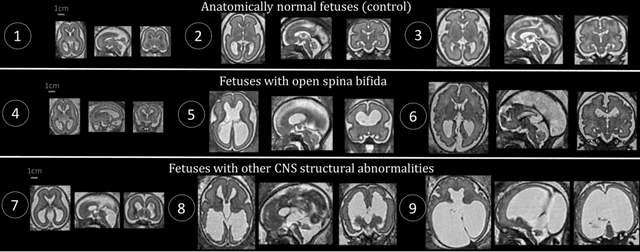
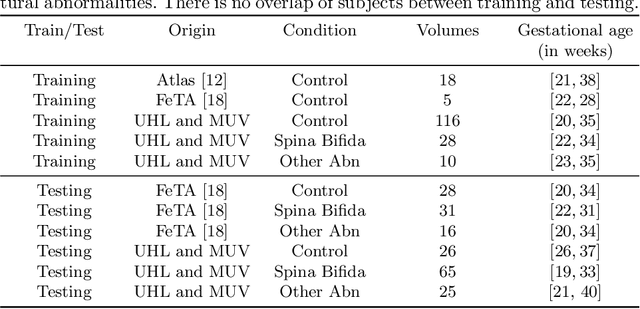
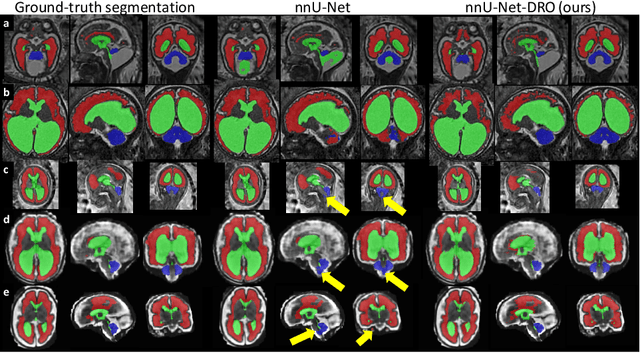
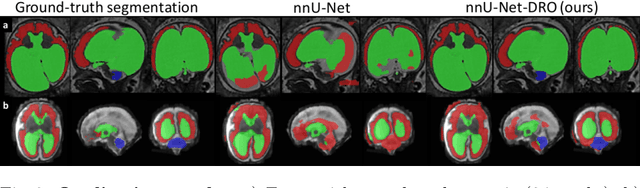
Abstract:The performance of deep neural networks typically increases with the number of training images. However, not all images have the same importance towards improved performance and robustness. In fetal brain MRI, abnormalities exacerbate the variability of the developing brain anatomy compared to non-pathological cases. A small number of abnormal cases, as is typically available in clinical datasets used for training, are unlikely to fairly represent the rich variability of abnormal developing brains. This leads machine learning systems trained by maximizing the average performance to be biased toward non-pathological cases. This problem was recently referred to as hidden stratification. To be suited for clinical use, automatic segmentation methods need to reliably achieve high-quality segmentation outcomes also for pathological cases. In this paper, we show that the state-of-the-art deep learning pipeline nnU-Net has difficulties to generalize to unseen abnormal cases. To mitigate this problem, we propose to train a deep neural network to minimize a percentile of the distribution of per-volume loss over the dataset. We show that this can be achieved by using Distributionally Robust Optimization (DRO). DRO automatically reweights the training samples with lower performance, encouraging nnU-Net to perform more consistently on all cases. We validated our approach using a dataset of 368 fetal brain T2w MRIs, including 124 MRIs of open spina bifida cases and 51 MRIs of cases with other severe abnormalities of brain development.
Label-set Loss Functions for Partial Supervision: Application to Fetal Brain 3D MRI Parcellation
Jul 09, 2021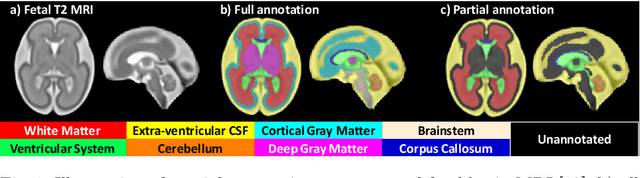
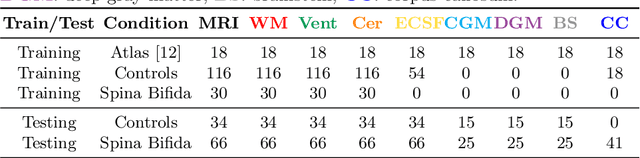
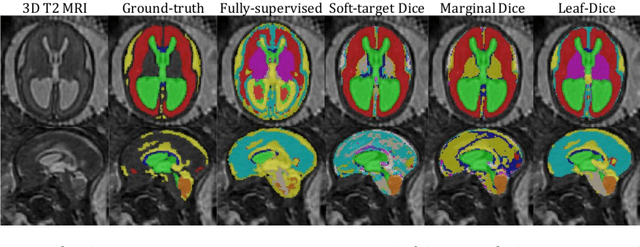
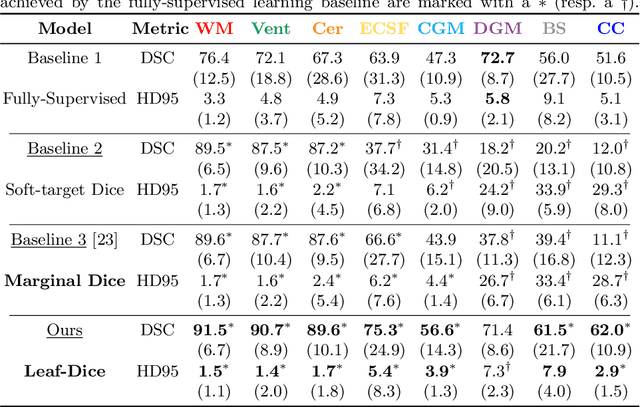
Abstract:Deep neural networks have increased the accuracy of automatic segmentation, however, their accuracy depends on the availability of a large number of fully segmented images. Methods to train deep neural networks using images for which some, but not all, regions of interest are segmented are necessary to make better use of partially annotated datasets. In this paper, we propose the first axiomatic definition of label-set loss functions that are the loss functions that can handle partially segmented images. We prove that there is one and only one method to convert a classical loss function for fully segmented images into a proper label-set loss function. Our theory also allows us to define the leaf-Dice loss, a label-set generalization of the Dice loss particularly suited for partial supervision with only missing labels. Using the leaf-Dice loss, we set a new state of the art in partially supervised learning for fetal brain 3D MRI segmentation. We achieve a deep neural network able to segment white matter, ventricles, cerebellum, extra-ventricular CSF, cortical gray matter, deep gray matter, brainstem, and corpus callosum based on fetal brain 3D MRI of anatomically normal fetuses or with open spina bifida. Our implementation of the proposed label-set loss functions is available at https://github.com/LucasFidon/label-set-loss-functions
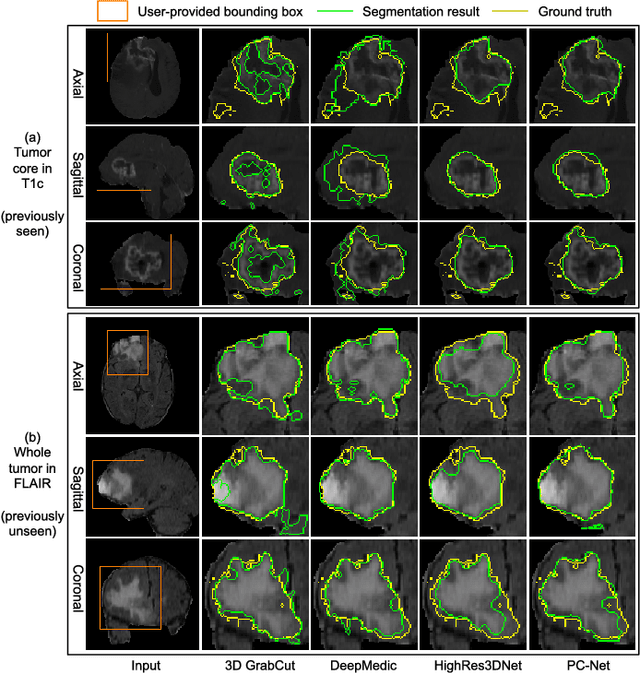
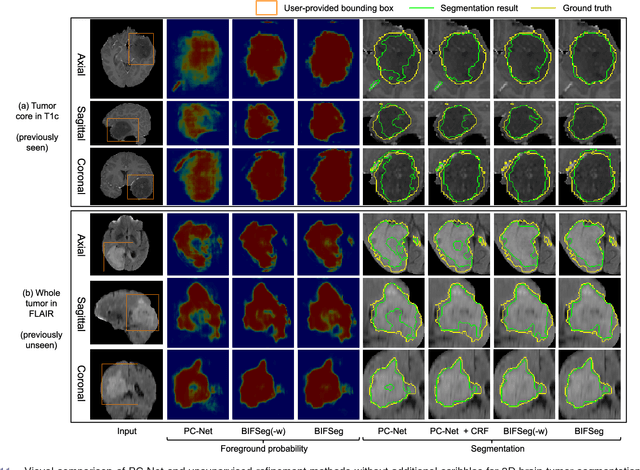
MIDeepSeg: Minimally Interactive Segmentation of Unseen Objects from Medical Images Using Deep Learning
Apr 25, 2021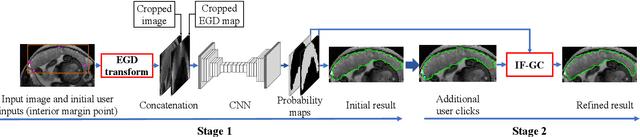
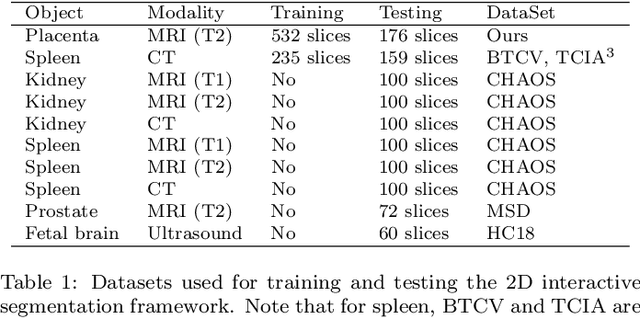
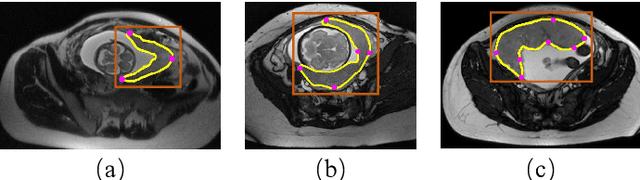
Abstract:Segmentation of organs or lesions from medical images plays an essential role in many clinical applications such as diagnosis and treatment planning. Though Convolutional Neural Networks (CNN) have achieved the state-of-the-art performance for automatic segmentation, they are often limited by the lack of clinically acceptable accuracy and robustness in complex cases. Therefore, interactive segmentation is a practical alternative to these methods. However, traditional interactive segmentation methods require a large amount of user interactions, and recently proposed CNN-based interactive segmentation methods are limited by poor performance on previously unseen objects. To solve these problems, we propose a novel deep learning-based interactive segmentation method that not only has high efficiency due to only requiring clicks as user inputs but also generalizes well to a range of previously unseen objects. Specifically, we first encode user-provided interior margin points via our proposed exponentialized geodesic distance that enables a CNN to achieve a good initial segmentation result of both previously seen and unseen objects, then we use a novel information fusion method that combines the initial segmentation with only few additional user clicks to efficiently obtain a refined segmentation. We validated our proposed framework through extensive experiments on 2D and 3D medical image segmentation tasks with a wide range of previous unseen objects that were not present in the training set. Experimental results showed that our proposed framework 1) achieves accurate results with fewer user interactions and less time compared with state-of-the-art interactive frameworks and 2) generalizes well to previously unseen objects.
CA-Net: Comprehensive Attention Convolutional Neural Networks for Explainable Medical Image Segmentation
Sep 23, 2020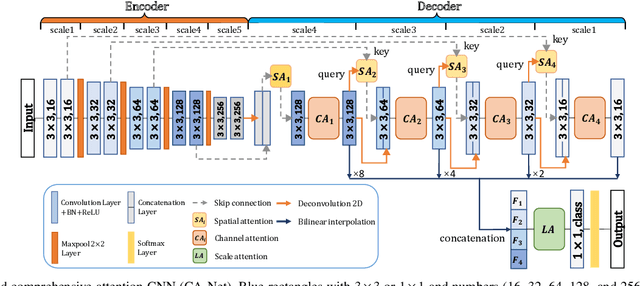
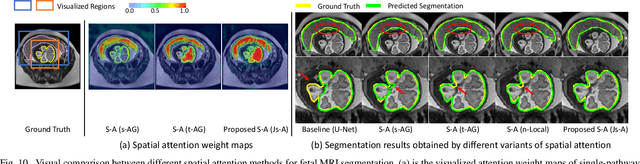
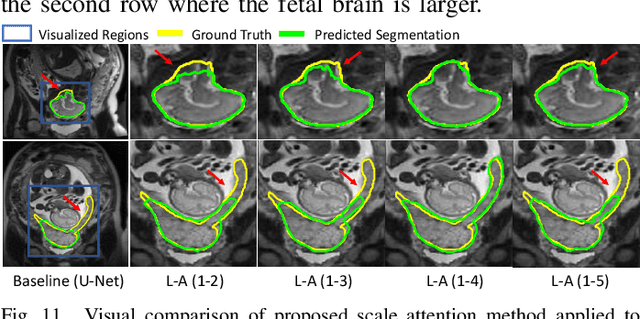
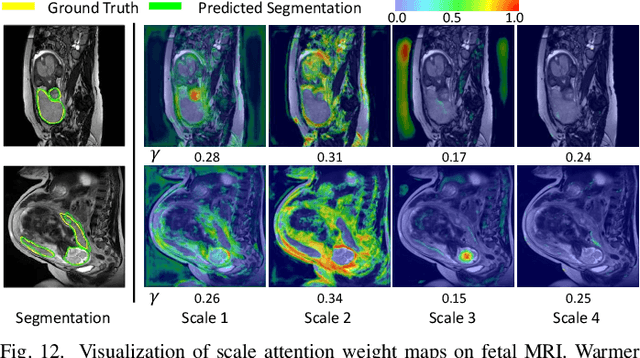
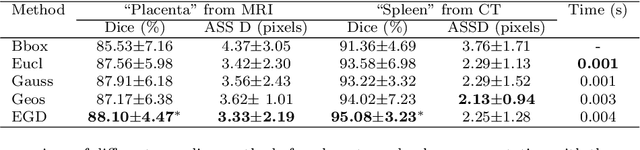
Abstract:Accurate medical image segmentation is essential for diagnosis and treatment planning of diseases. Convolutional Neural Networks (CNNs) have achieved state-of-the-art performance for automatic medical image segmentation. However, they are still challenged by complicated conditions where the segmentation target has large variations of position, shape and scale, and existing CNNs have a poor explainability that limits their application to clinical decisions. In this work, we make extensive use of multiple attentions in a CNN architecture and propose a comprehensive attention-based CNN (CA-Net) for more accurate and explainable medical image segmentation that is aware of the most important spatial positions, channels and scales at the same time. In particular, we first propose a joint spatial attention module to make the network focus more on the foreground region. Then, a novel channel attention module is proposed to adaptively recalibrate channel-wise feature responses and highlight the most relevant feature channels. Also, we propose a scale attention module implicitly emphasizing the most salient feature maps among multiple scales so that the CNN is adaptive to the size of an object. Extensive experiments on skin lesion segmentation from ISIC 2018 and multi-class segmentation of fetal MRI found that our proposed CA-Net significantly improved the average segmentation Dice score from 87.77% to 92.08% for skin lesion, 84.79% to 87.08% for the placenta and 93.20% to 95.88% for the fetal brain respectively compared with U-Net. It reduced the model size to around 15 times smaller with close or even better accuracy compared with state-of-the-art DeepLabv3+. In addition, it has a much higher explainability than existing networks by visualizing the attention weight maps. Our code is available at https://github.com/HiLab-git/CA-Net
Uncertainty-Guided Efficient Interactive Refinement of Fetal Brain Segmentation from Stacks of MRI Slices
Jul 02, 2020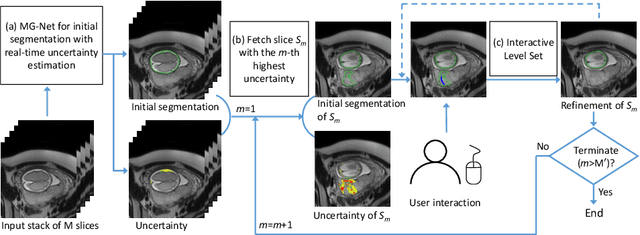
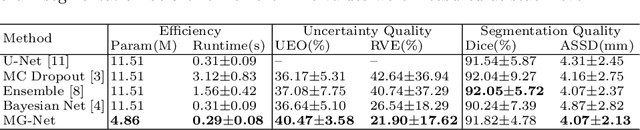
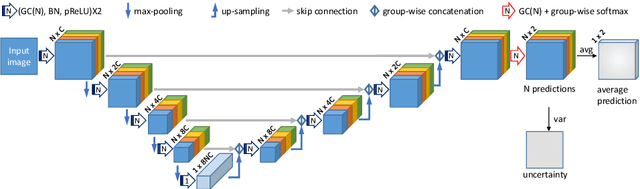
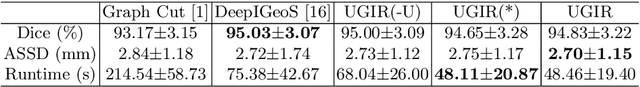
Abstract:Segmentation of the fetal brain from stacks of motion-corrupted fetal MRI slices is important for motion correction and high-resolution volume reconstruction. Although Convolutional Neural Networks (CNNs) have been widely used for automatic segmentation of the fetal brain, their results may still benefit from interactive refinement for challenging slices. To improve the efficiency of interactive refinement process, we propose an Uncertainty-Guided Interactive Refinement (UGIR) framework. We first propose a grouped convolution-based CNN to obtain multiple automatic segmentation predictions with uncertainty estimation in a single forward pass, then guide the user to provide interactions only in a subset of slices with the highest uncertainty. A novel interactive level set method is also proposed to obtain a refined result given the initial segmentation and user interactions. Experimental results show that: (1) our proposed CNN obtains uncertainty estimation in real time which correlates well with mis-segmentations, (2) the proposed interactive level set is effective and efficient for refinement, (3) UGIR obtains accurate refinement results with around 30% improvement of efficiency by using uncertainty to guide user interactions. Our code is available online.
Aleatoric uncertainty estimation with test-time augmentation for medical image segmentation with convolutional neural networks
Jul 20, 2018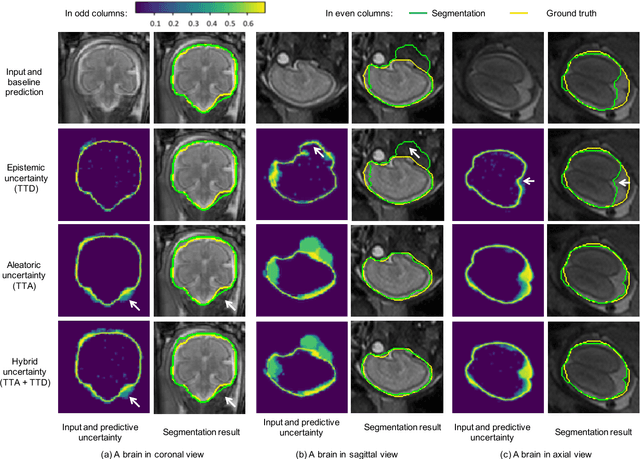
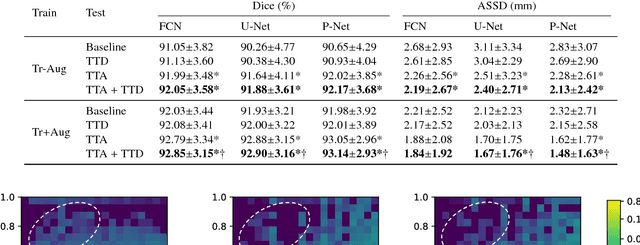
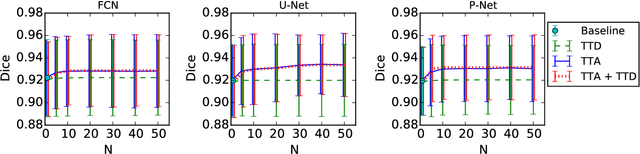
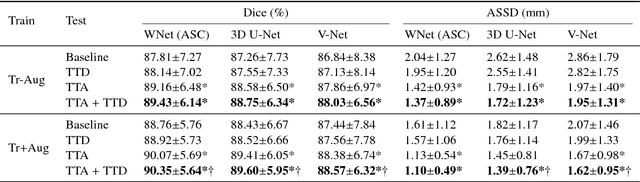
Abstract:Despite the state-of-the-art performance for medical image segmentation, deep convolutional neural networks (CNNs) have rarely provided uncertainty estimations regarding their segmentation outputs, e.g., model (epistemic) and image-based (aleatoric) uncertainties. In this work, we analyze these different types of uncertainties for CNN-based 2D and 3D medical image segmentation tasks. We additionally propose a test-time augmentation-based aleatoric uncertainty to analyze the effect of different transformations of the input image on the segmentation output. Test-time augmentation has been previously used to improve segmentation accuracy, yet not been formulated in a consistent mathematical framework. Hence, we also propose a theoretical formulation of test-time augmentation, where a distribution of the prediction is estimated by Monte Carlo simulation with prior distributions of parameters in an image acquisition model that involves image transformations and noise. We compare and combine our proposed aleatoric uncertainty with model uncertainty. Experiments with segmentation of fetal brains and brain tumors from 2D and 3D Magnetic Resonance Images (MRI) showed that 1) the test-time augmentation-based aleatoric uncertainty provides a better uncertainty estimation than calculating the test-time dropout-based model uncertainty alone and helps to reduce overconfident incorrect predictions, and 2) our test-time augmentation outperforms a single-prediction baseline and dropout-based multiple predictions.
Interactive Medical Image Segmentation using Deep Learning with Image-specific Fine-tuning
Oct 11, 2017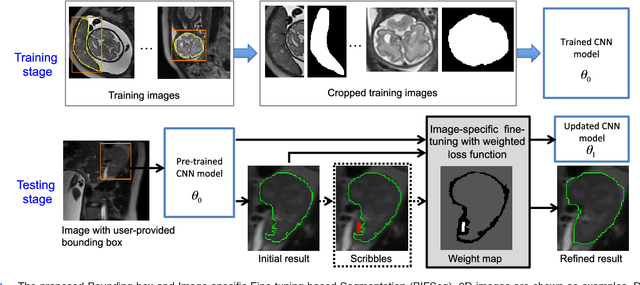
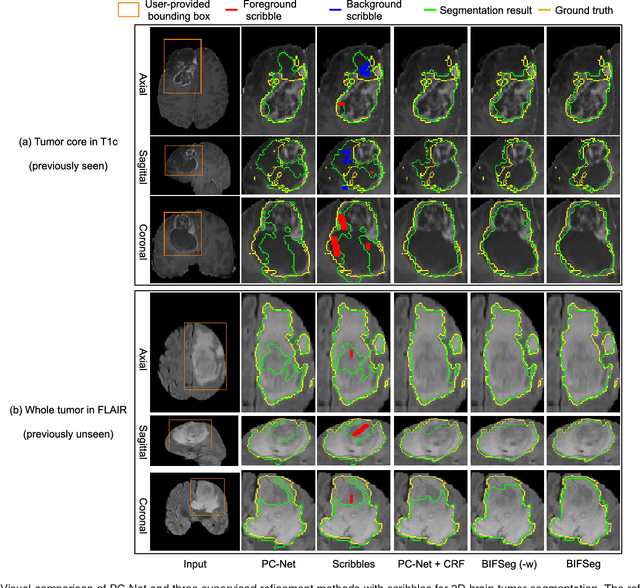
Abstract:Convolutional neural networks (CNNs) have achieved state-of-the-art performance for automatic medical image segmentation. However, they have not demonstrated sufficiently accurate and robust results for clinical use. In addition, they are limited by the lack of image-specific adaptation and the lack of generalizability to previously unseen object classes. To address these problems, we propose a novel deep learning-based framework for interactive segmentation by incorporating CNNs into a bounding box and scribble-based segmentation pipeline. We propose image-specific fine-tuning to make a CNN model adaptive to a specific test image, which can be either unsupervised (without additional user interactions) or supervised (with additional scribbles). We also propose a weighted loss function considering network and interaction-based uncertainty for the fine-tuning. We applied this framework to two applications: 2D segmentation of multiple organs from fetal MR slices, where only two types of these organs were annotated for training; and 3D segmentation of brain tumor core (excluding edema) and whole brain tumor (including edema) from different MR sequences, where only tumor cores in one MR sequence were annotated for training. Experimental results show that 1) our model is more robust to segment previously unseen objects than state-of-the-art CNNs; 2) image-specific fine-tuning with the proposed weighted loss function significantly improves segmentation accuracy; and 3) our method leads to accurate results with fewer user interactions and less user time than traditional interactive segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge