András Jakab
Brain Tumor Segmentation (BraTS) Challenge 2024: Meningioma Radiotherapy Planning Automated Segmentation
May 28, 2024Abstract:The 2024 Brain Tumor Segmentation Meningioma Radiotherapy (BraTS-MEN-RT) challenge aims to advance automated segmentation algorithms using the largest known multi-institutional dataset of radiotherapy planning brain MRIs with expert-annotated target labels for patients with intact or post-operative meningioma that underwent either conventional external beam radiotherapy or stereotactic radiosurgery. Each case includes a defaced 3D post-contrast T1-weighted radiotherapy planning MRI in its native acquisition space, accompanied by a single-label "target volume" representing the gross tumor volume (GTV) and any at-risk post-operative site. Target volume annotations adhere to established radiotherapy planning protocols, ensuring consistency across cases and institutions. For pre-operative meningiomas, the target volume encompasses the entire GTV and associated nodular dural tail, while for post-operative cases, it includes at-risk resection cavity margins as determined by the treating institution. Case annotations were reviewed and approved by expert neuroradiologists and radiation oncologists. Participating teams will develop, containerize, and evaluate automated segmentation models using this comprehensive dataset. Model performance will be assessed using the lesion-wise Dice Similarity Coefficient and the 95% Hausdorff distance. The top-performing teams will be recognized at the Medical Image Computing and Computer Assisted Intervention Conference in October 2024. BraTS-MEN-RT is expected to significantly advance automated radiotherapy planning by enabling precise tumor segmentation and facilitating tailored treatment, ultimately improving patient outcomes.
Analysis of the BraTS 2023 Intracranial Meningioma Segmentation Challenge
May 16, 2024



Abstract:We describe the design and results from the BraTS 2023 Intracranial Meningioma Segmentation Challenge. The BraTS Meningioma Challenge differed from prior BraTS Glioma challenges in that it focused on meningiomas, which are typically benign extra-axial tumors with diverse radiologic and anatomical presentation and a propensity for multiplicity. Nine participating teams each developed deep-learning automated segmentation models using image data from the largest multi-institutional systematically expert annotated multilabel multi-sequence meningioma MRI dataset to date, which included 1000 training set cases, 141 validation set cases, and 283 hidden test set cases. Each case included T2, T2/FLAIR, T1, and T1Gd brain MRI sequences with associated tumor compartment labels delineating enhancing tumor, non-enhancing tumor, and surrounding non-enhancing T2/FLAIR hyperintensity. Participant automated segmentation models were evaluated and ranked based on a scoring system evaluating lesion-wise metrics including dice similarity coefficient (DSC) and 95% Hausdorff Distance. The top ranked team had a lesion-wise median dice similarity coefficient (DSC) of 0.976, 0.976, and 0.964 for enhancing tumor, tumor core, and whole tumor, respectively and a corresponding average DSC of 0.899, 0.904, and 0.871, respectively. These results serve as state-of-the-art benchmarks for future pre-operative meningioma automated segmentation algorithms. Additionally, we found that 1286 of 1424 cases (90.3%) had at least 1 compartment voxel abutting the edge of the skull-stripped image edge, which requires further investigation into optimal pre-processing face anonymization steps.
A Dempster-Shafer approach to trustworthy AI with application to fetal brain MRI segmentation
Apr 05, 2022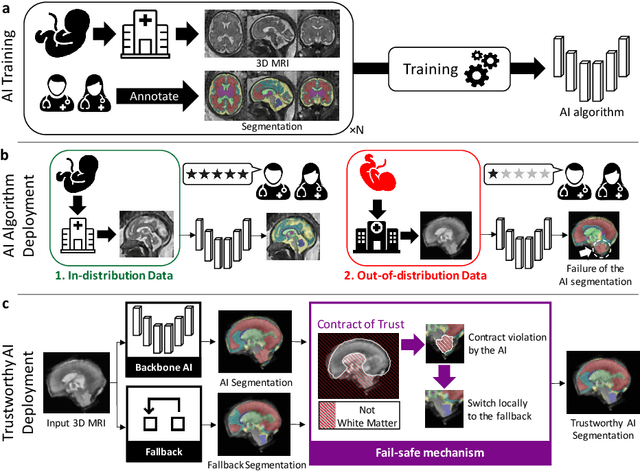
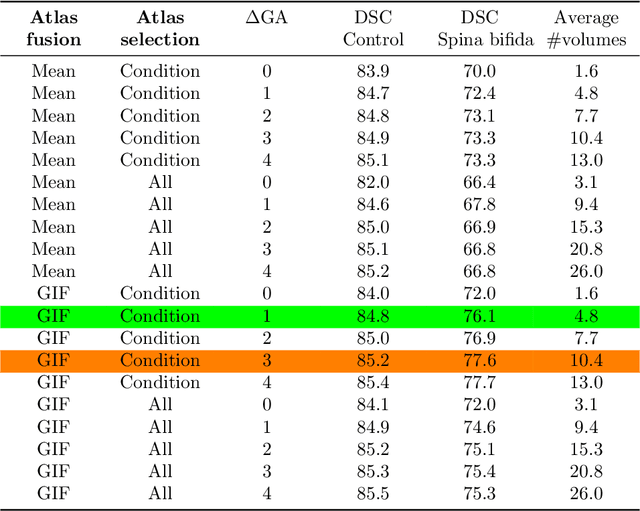
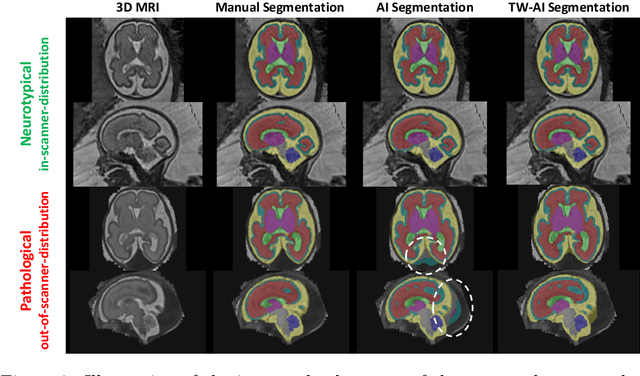
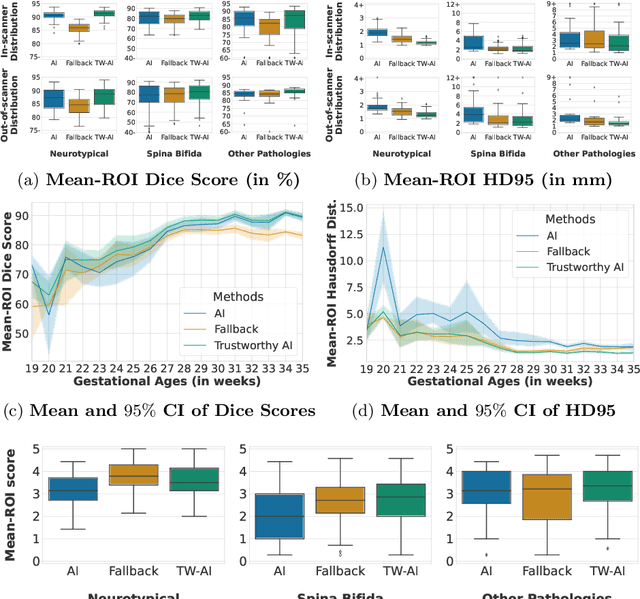
Abstract:Deep learning models for medical image segmentation can fail unexpectedly and spectacularly for pathological cases and for images acquired at different centers than those used for training, with labeling errors that violate expert knowledge about the anatomy and the intensity distribution of the regions to be segmented. Such errors undermine the trustworthiness of deep learning models developed for medical image segmentation. Mechanisms with a fallback method for detecting and correcting such failures are essential for safely translating this technology into clinics and are likely to be a requirement of future regulations on artificial intelligence (AI). Here, we propose a principled trustworthy AI theoretical framework and a practical system that can augment any backbone AI system using a fallback method and a fail-safe mechanism based on Dempster-Shafer theory. Our approach relies on an actionable definition of trustworthy AI. Our method automatically discards the voxel-level labeling predicted by the backbone AI that are likely to violate expert knowledge and relies on a fallback atlas-based segmentation method for those voxels. We demonstrate the effectiveness of the proposed trustworthy AI approach on the largest reported annotated dataset of fetal T2w MRI consisting of 540 manually annotated fetal brain 3D MRIs with neurotypical or abnormal brain development and acquired from 13 sources of data across 6 countries. We show that our trustworthy AI method improves the robustness of a state-of-the-art backbone AI for fetal brain MRI segmentation on MRIs acquired across various centers and for fetuses with various brain abnormalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge