Gregor Kasprian
Conditional Fetal Brain Atlas Learning for Automatic Tissue Segmentation
Aug 06, 2025Abstract:Magnetic Resonance Imaging (MRI) of the fetal brain has become a key tool for studying brain development in vivo. Yet, its assessment remains challenging due to variability in brain maturation, imaging protocols, and uncertain estimates of Gestational Age (GA). To overcome these, brain atlases provide a standardized reference framework that facilitates objective evaluation and comparison across subjects by aligning the atlas and subjects in a common coordinate system. In this work, we introduce a novel deep-learning framework for generating continuous, age-specific fetal brain atlases for real-time fetal brain tissue segmentation. The framework combines a direct registration model with a conditional discriminator. Trained on a curated dataset of 219 neurotypical fetal MRIs spanning from 21 to 37 weeks of gestation. The method achieves high registration accuracy, captures dynamic anatomical changes with sharp structural detail, and robust segmentation performance with an average Dice Similarity Coefficient (DSC) of 86.3% across six brain tissues. Furthermore, volumetric analysis of the generated atlases reveals detailed neurotypical growth trajectories, providing valuable insights into the maturation of the fetal brain. This approach enables individualized developmental assessment with minimal pre-processing and real-time performance, supporting both research and clinical applications. The model code is available at https://github.com/cirmuw/fetal-brain-atlas
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025



Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Towards contrast- and pathology-agnostic clinical fetal brain MRI segmentation using SynthSeg
Apr 14, 2025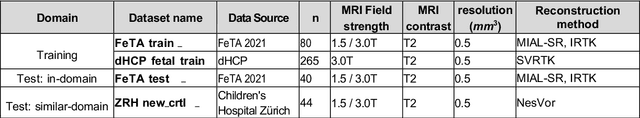



Abstract:Magnetic resonance imaging (MRI) has played a crucial role in fetal neurodevelopmental research. Structural annotations of MR images are an important step for quantitative analysis of the developing human brain, with Deep learning providing an automated alternative for this otherwise tedious manual process. However, segmentation performances of Convolutional Neural Networks often suffer from domain shift, where the network fails when applied to subjects that deviate from the distribution with which it is trained on. In this work, we aim to train networks capable of automatically segmenting fetal brain MRIs with a wide range of domain shifts pertaining to differences in subject physiology and acquisition environments, in particular shape-based differences commonly observed in pathological cases. We introduce a novel data-driven train-time sampling strategy that seeks to fully exploit the diversity of a given training dataset to enhance the domain generalizability of the trained networks. We adapted our sampler, together with other existing data augmentation techniques, to the SynthSeg framework, a generator that utilizes domain randomization to generate diverse training data, and ran thorough experimentations and ablation studies on a wide range of training/testing data to test the validity of the approaches. Our networks achieved notable improvements in the segmentation quality on testing subjects with intense anatomical abnormalities (p < 1e-4), though at the cost of a slighter decrease in performance in cases with fewer abnormalities. Our work also lays the foundation for future works on creating and adapting data-driven sampling strategies for other training pipelines.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024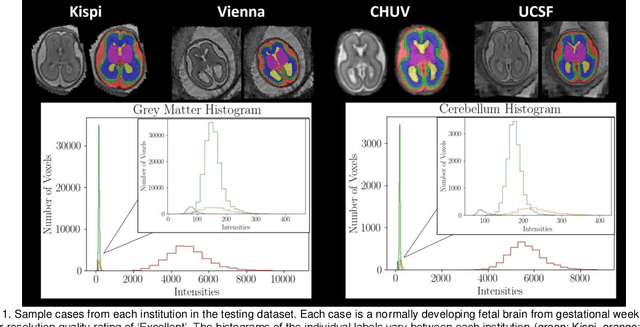
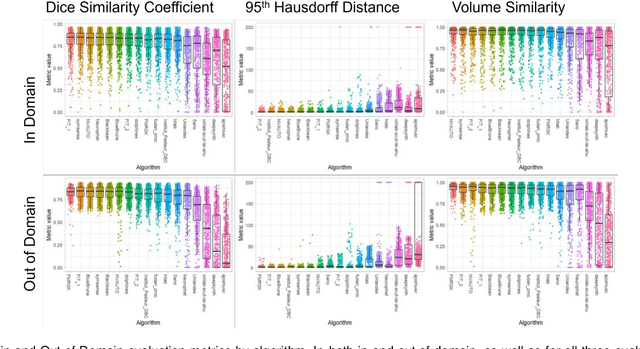
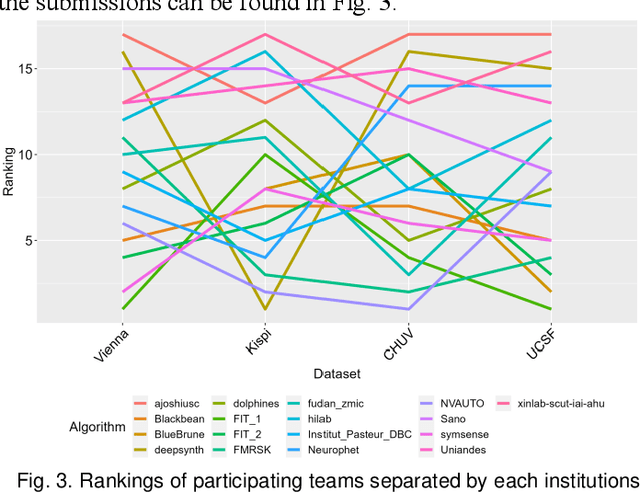
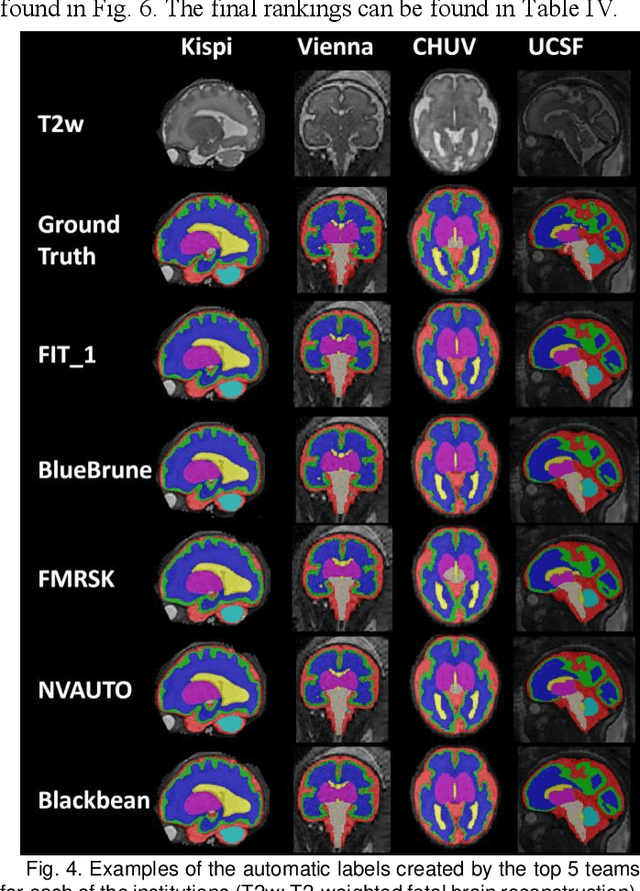
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
Spatio-temporal motion correction and iterative reconstruction of in-utero fetal fMRI
Sep 17, 2022Abstract:Resting-state functional Magnetic Resonance Imaging (fMRI) is a powerful imaging technique for studying functional development of the brain in utero. However, unpredictable and excessive movement of fetuses have limited its clinical applicability. Previous studies have focused primarily on the accurate estimation of the motion parameters employing a single step 3D interpolation at each individual time frame to recover a motion-free 4D fMRI image. Using only information from a 3D spatial neighborhood neglects the temporal structure of fMRI and useful information from neighboring timepoints. Here, we propose a novel technique based on four dimensional iterative reconstruction of the motion scattered fMRI slices. Quantitative evaluation of the proposed method on a cohort of real clinical fetal fMRI data indicates improvement of reconstruction quality compared to the conventional 3D interpolation approaches.
A Dempster-Shafer approach to trustworthy AI with application to fetal brain MRI segmentation
Apr 05, 2022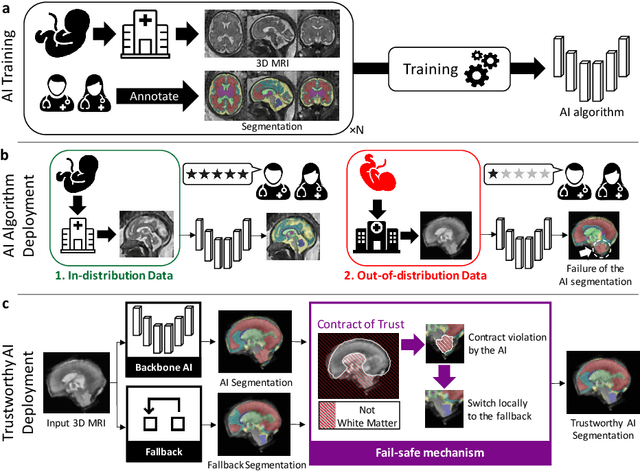
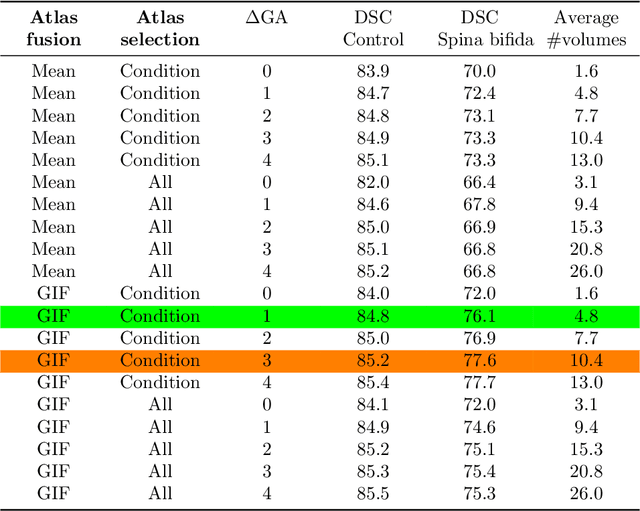
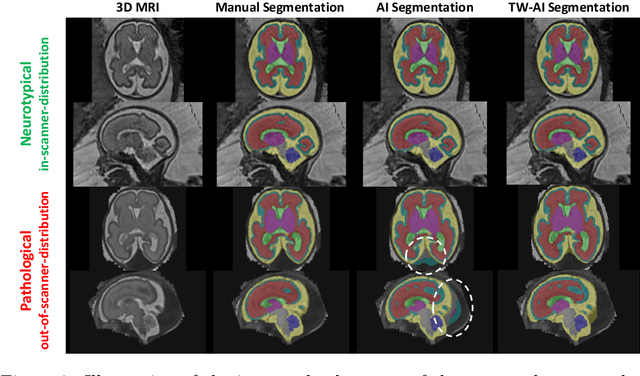
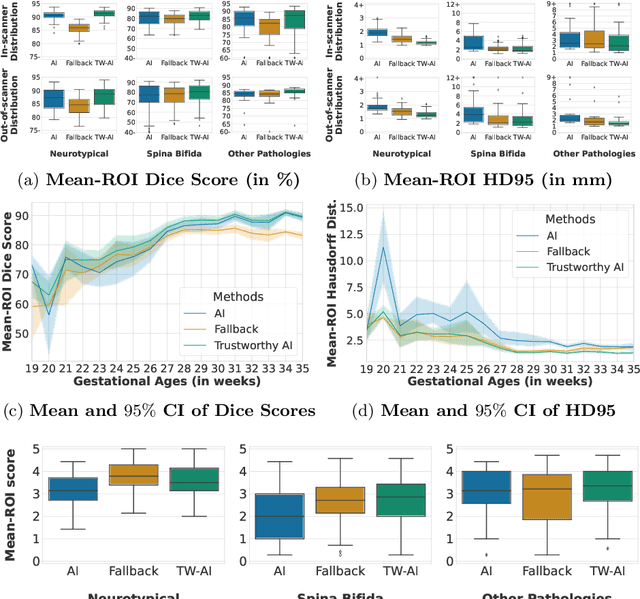
Abstract:Deep learning models for medical image segmentation can fail unexpectedly and spectacularly for pathological cases and for images acquired at different centers than those used for training, with labeling errors that violate expert knowledge about the anatomy and the intensity distribution of the regions to be segmented. Such errors undermine the trustworthiness of deep learning models developed for medical image segmentation. Mechanisms with a fallback method for detecting and correcting such failures are essential for safely translating this technology into clinics and are likely to be a requirement of future regulations on artificial intelligence (AI). Here, we propose a principled trustworthy AI theoretical framework and a practical system that can augment any backbone AI system using a fallback method and a fail-safe mechanism based on Dempster-Shafer theory. Our approach relies on an actionable definition of trustworthy AI. Our method automatically discards the voxel-level labeling predicted by the backbone AI that are likely to violate expert knowledge and relies on a fallback atlas-based segmentation method for those voxels. We demonstrate the effectiveness of the proposed trustworthy AI approach on the largest reported annotated dataset of fetal T2w MRI consisting of 540 manually annotated fetal brain 3D MRIs with neurotypical or abnormal brain development and acquired from 13 sources of data across 6 countries. We show that our trustworthy AI method improves the robustness of a state-of-the-art backbone AI for fetal brain MRI segmentation on MRIs acquired across various centers and for fetuses with various brain abnormalities.
Motion Correction and Volumetric Reconstruction for Fetal Functional Magnetic Resonance Imaging Data
Feb 11, 2022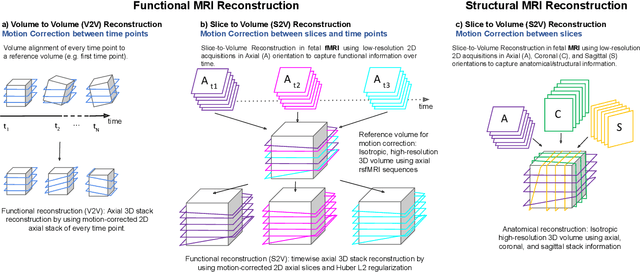
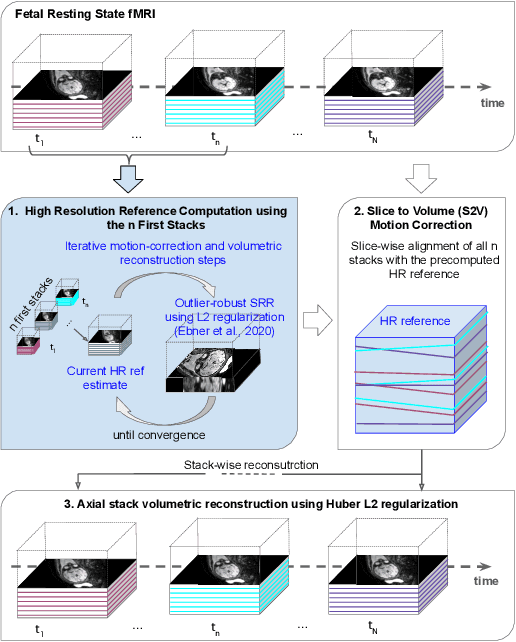
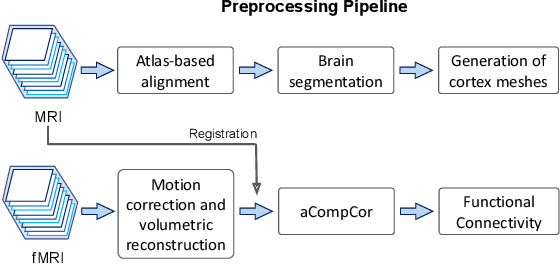
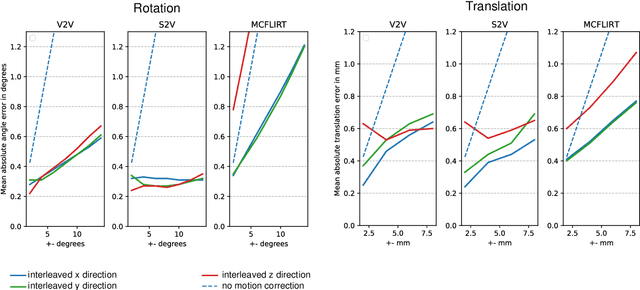
Abstract:Motion correction is an essential preprocessing step in functional Magnetic Resonance Imaging (fMRI) of the fetal brain with the aim to remove artifacts caused by fetal movement and maternal breathing and consequently to suppress erroneous signal correlations. Current motion correction approaches for fetal fMRI choose a single 3D volume from a specific acquisition timepoint with least motion artefacts as reference volume, and perform interpolation for the reconstruction of the motion corrected time series. The results can suffer, if no low-motion frame is available, and if reconstruction does not exploit any assumptions about the continuity of the fMRI signal. Here, we propose a novel framework, which estimates a high-resolution reference volume by using outlier-robust motion correction, and by utilizing Huber L2 regularization for intra-stack volumetric reconstruction of the motion-corrected fetal brain fMRI. We performed an extensive parameter study to investigate the effectiveness of motion estimation and present in this work benchmark metrics to quantify the effect of motion correction and regularised volumetric reconstruction approaches on functional connectivity computations. We demonstrate the proposed framework's ability to improve functional connectivity estimates, reproducibility and signal interpretability, which is clinically highly desirable for the establishment of prognostic noninvasive imaging biomarkers. The motion correction and volumetric reconstruction framework is made available as an open-source package of NiftyMIC.
Distributionally Robust Segmentation of Abnormal Fetal Brain 3D MRI
Aug 09, 2021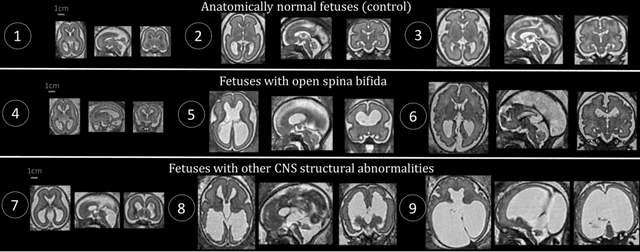
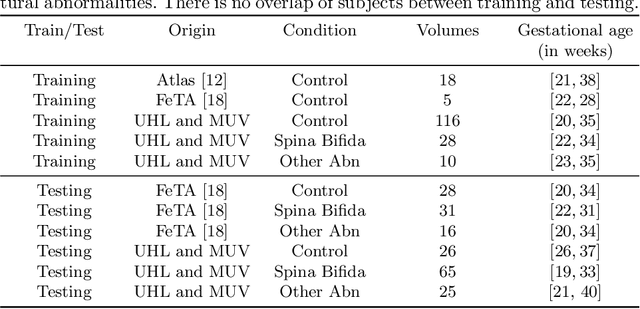
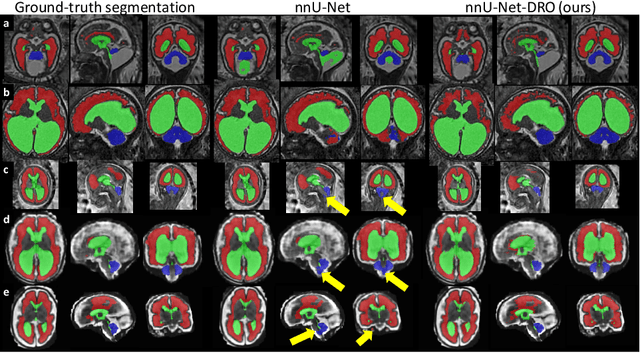
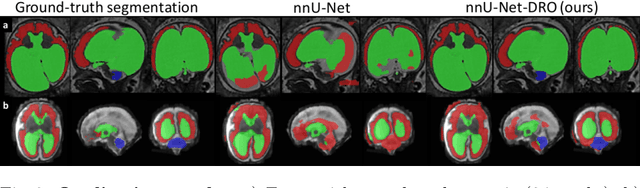
Abstract:The performance of deep neural networks typically increases with the number of training images. However, not all images have the same importance towards improved performance and robustness. In fetal brain MRI, abnormalities exacerbate the variability of the developing brain anatomy compared to non-pathological cases. A small number of abnormal cases, as is typically available in clinical datasets used for training, are unlikely to fairly represent the rich variability of abnormal developing brains. This leads machine learning systems trained by maximizing the average performance to be biased toward non-pathological cases. This problem was recently referred to as hidden stratification. To be suited for clinical use, automatic segmentation methods need to reliably achieve high-quality segmentation outcomes also for pathological cases. In this paper, we show that the state-of-the-art deep learning pipeline nnU-Net has difficulties to generalize to unseen abnormal cases. To mitigate this problem, we propose to train a deep neural network to minimize a percentile of the distribution of per-volume loss over the dataset. We show that this can be achieved by using Distributionally Robust Optimization (DRO). DRO automatically reweights the training samples with lower performance, encouraging nnU-Net to perform more consistently on all cases. We validated our approach using a dataset of 368 fetal brain T2w MRIs, including 124 MRIs of open spina bifida cases and 51 MRIs of cases with other severe abnormalities of brain development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge