Lang Li
ClinicalTrialsHub: Bridging Registries and Literature for Comprehensive Clinical Trial Access
Dec 09, 2025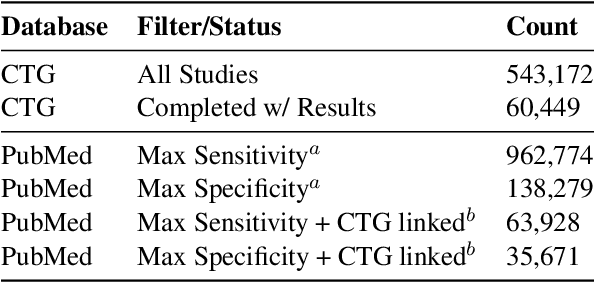
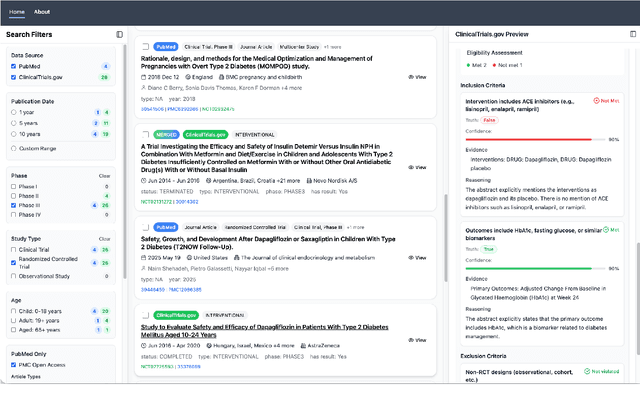
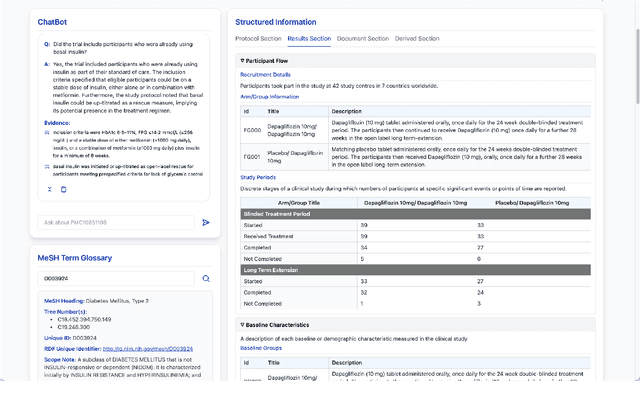
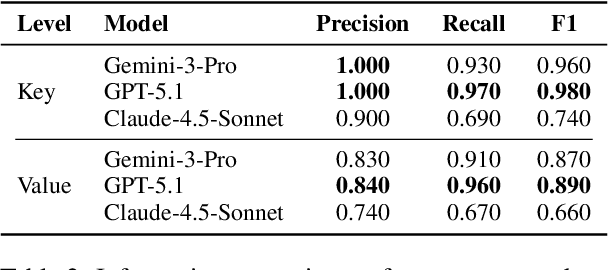
Abstract:We present ClinicalTrialsHub, an interactive search-focused platform that consolidates all data from ClinicalTrials.gov and augments it by automatically extracting and structuring trial-relevant information from PubMed research articles. Our system effectively increases access to structured clinical trial data by 83.8% compared to relying on ClinicalTrials.gov alone, with potential to make access easier for patients, clinicians, researchers, and policymakers, advancing evidence-based medicine. ClinicalTrialsHub uses large language models such as GPT-5.1 and Gemini-3-Pro to enhance accessibility. The platform automatically parses full-text research articles to extract structured trial information, translates user queries into structured database searches, and provides an attributed question-answering system that generates evidence-grounded answers linked to specific source sentences. We demonstrate its utility through a user study involving clinicians, clinical researchers, and PhD students of pharmaceutical sciences and nursing, and a systematic automatic evaluation of its information extraction and question answering capabilities.
Ontology- and LLM-based Data Harmonization for Federated Learning in Healthcare
May 26, 2025Abstract:The rise of electronic health records (EHRs) has unlocked new opportunities for medical research, but privacy regulations and data heterogeneity remain key barriers to large-scale machine learning. Federated learning (FL) enables collaborative modeling without sharing raw data, yet faces challenges in harmonizing diverse clinical datasets. This paper presents a two-step data alignment strategy integrating ontologies and large language models (LLMs) to support secure, privacy-preserving FL in healthcare, demonstrating its effectiveness in a real-world project involving semantic mapping of EHR data.
MultiChartQA: Benchmarking Vision-Language Models on Multi-Chart Problems
Oct 18, 2024



Abstract:Multimodal Large Language Models (MLLMs) have demonstrated impressive abilities across various tasks, including visual question answering and chart comprehension, yet existing benchmarks for chart-related tasks fall short in capturing the complexity of real-world multi-chart scenarios. Current benchmarks primarily focus on single-chart tasks, neglecting the multi-hop reasoning required to extract and integrate information from multiple charts, which is essential in practical applications. To fill this gap, we introduce MultiChartQA, a benchmark that evaluates MLLMs' capabilities in four key areas: direct question answering, parallel question answering, comparative reasoning, and sequential reasoning. Our evaluation of a wide range of MLLMs reveals significant performance gaps compared to humans. These results highlight the challenges in multi-chart comprehension and the potential of MultiChartQA to drive advancements in this field. Our code and data are available at https://github.com/Zivenzhu/Multi-chart-QA
Training-Free Large Model Priors for Multiple-in-One Image Restoration
Jul 18, 2024Abstract:Image restoration aims to reconstruct the latent clear images from their degraded versions. Despite the notable achievement, existing methods predominantly focus on handling specific degradation types and thus require specialized models, impeding real-world applications in dynamic degradation scenarios. To address this issue, we propose Large Model Driven Image Restoration framework (LMDIR), a novel multiple-in-one image restoration paradigm that leverages the generic priors from large multi-modal language models (MMLMs) and the pretrained diffusion models. In detail, LMDIR integrates three key prior knowledges: 1) global degradation knowledge from MMLMs, 2) scene-aware contextual descriptions generated by MMLMs, and 3) fine-grained high-quality reference images synthesized by diffusion models guided by MMLM descriptions. Standing on above priors, our architecture comprises a query-based prompt encoder, degradation-aware transformer block injecting global degradation knowledge, content-aware transformer block incorporating scene description, and reference-based transformer block incorporating fine-grained image priors. This design facilitates single-stage training paradigm to address various degradations while supporting both automatic and user-guided restoration. Extensive experiments demonstrate that our designed method outperforms state-of-the-art competitors on multiple evaluation benchmarks.
SubgroupTE: Advancing Treatment Effect Estimation with Subgroup Identification
Jan 22, 2024
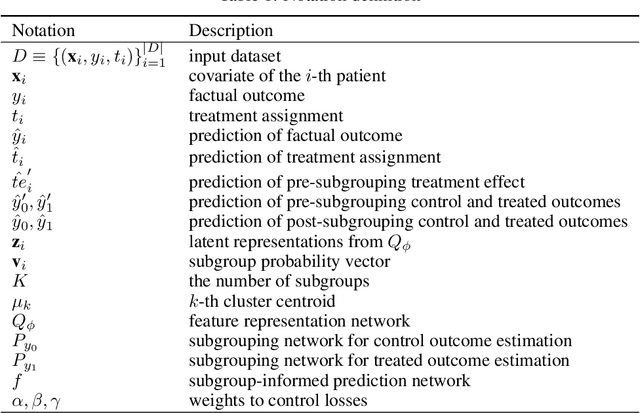
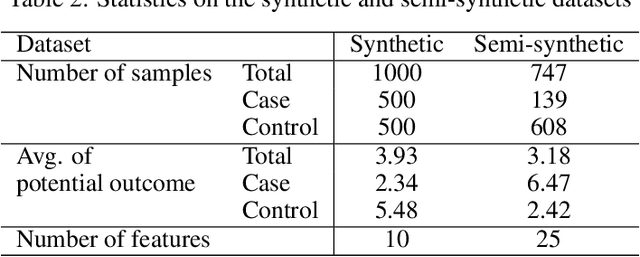
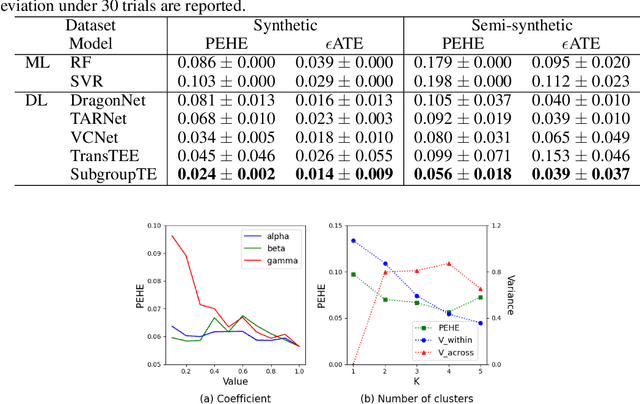
Abstract:Precise estimation of treatment effects is crucial for evaluating intervention effectiveness. While deep learning models have exhibited promising performance in learning counterfactual representations for treatment effect estimation (TEE), a major limitation in most of these models is that they treat the entire population as a homogeneous group, overlooking the diversity of treatment effects across potential subgroups that have varying treatment effects. This limitation restricts the ability to precisely estimate treatment effects and provide subgroup-specific treatment recommendations. In this paper, we propose a novel treatment effect estimation model, named SubgroupTE, which incorporates subgroup identification in TEE. SubgroupTE identifies heterogeneous subgroups with different treatment responses and more precisely estimates treatment effects by considering subgroup-specific causal effects. In addition, SubgroupTE iteratively optimizes subgrouping and treatment effect estimation networks to enhance both estimation and subgroup identification. Comprehensive experiments on the synthetic and semi-synthetic datasets exhibit the outstanding performance of SubgroupTE compared with the state-of-the-art models on treatment effect estimation. Additionally, a real-world study demonstrates the capabilities of SubgroupTE in enhancing personalized treatment recommendations for patients with opioid use disorder (OUD) by advancing treatment effect estimation with subgroup identification.
Thinking about GPT-3 In-Context Learning for Biomedical IE? Think Again
Mar 16, 2022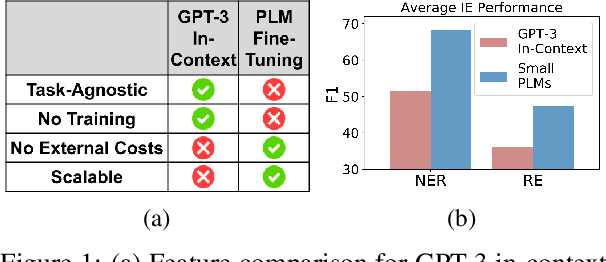
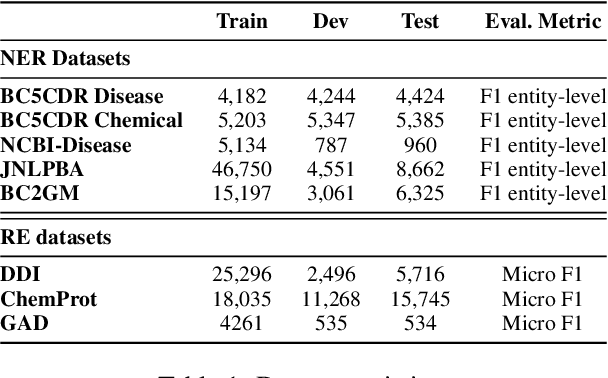

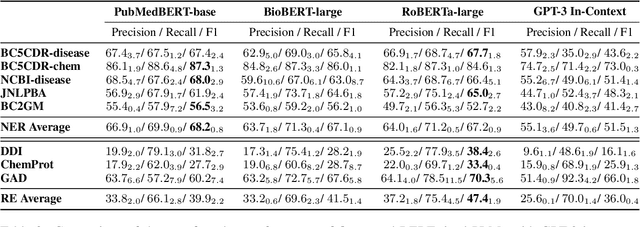
Abstract:The strong few-shot in-context learning capability of large pre-trained language models (PLMs) such as GPT-3 is highly appealing for biomedical applications where data annotation is particularly costly. In this paper, we present the first systematic and comprehensive study to compare the few-shot performance of GPT-3 in-context learning with fine-tuning smaller (i.e., BERT-sized) PLMs on two highly representative biomedical information extraction tasks, named entity recognition and relation extraction. We follow the true few-shot setting to avoid overestimating models' few-shot performance by model selection over a large validation set. We also optimize GPT-3's performance with known techniques such as contextual calibration and dynamic in-context example retrieval. However, our results show that GPT-3 still significantly underperforms compared with simply fine-tuning a smaller PLM using the same small training set. Moreover, what is equally important for practical applications is that adding more labeled data would reliably yield an improvement in model performance. While that is the case when fine-tuning small PLMs, GPT-3's performance barely improves when adding more data. In-depth analyses further reveal issues of the in-context learning setting that may be detrimental to information extraction tasks in general. Given the high cost of experimenting with GPT-3, we hope our study provides guidance for biomedical researchers and practitioners towards more promising directions such as fine-tuning GPT-3 or small PLMs.
Drug-drug interaction prediction based on co-medication patterns and graph matching
Feb 22, 2019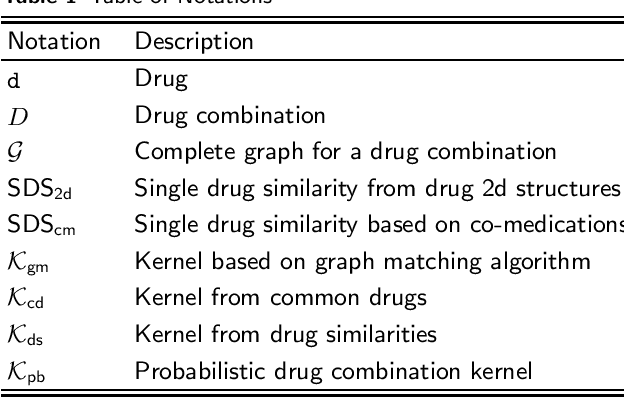
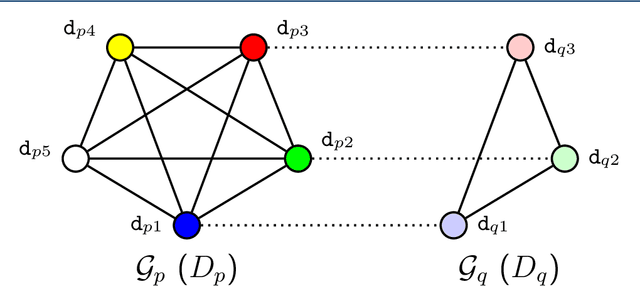
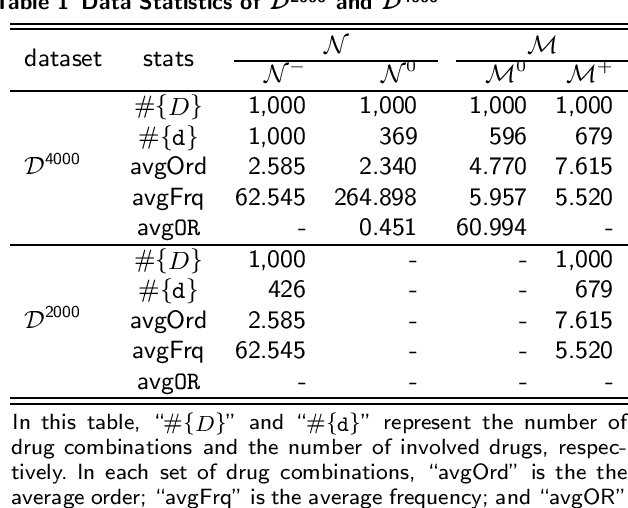
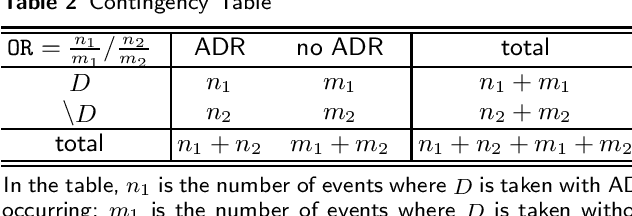
Abstract:Background: The problem of predicting whether a drug combination of arbitrary orders is likely to induce adverse drug reactions is considered in this manuscript. Methods: Novel kernels over drug combinations of arbitrary orders are developed within support vector machines for the prediction. Graph matching methods are used in the novel kernels to measure the similarities among drug combinations, in which drug co-medication patterns are leveraged to measure single drug similarities. Results: The experimental results on a real-world dataset demonstrated that the new kernels achieve an area under the curve (AUC) value 0.912 for the prediction problem. Conclusions: The new methods with drug co-medication based single drug similarities can accurately predict whether a drug combination is likely to induce adverse drug reactions of interest. Keywords: drug-drug interaction prediction; drug combination similarity; co-medication; graph matching
Drug Recommendation toward Safe Polypharmacy
Mar 08, 2018
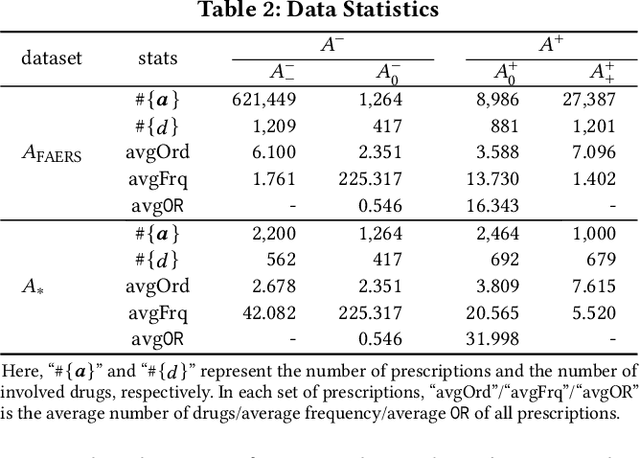
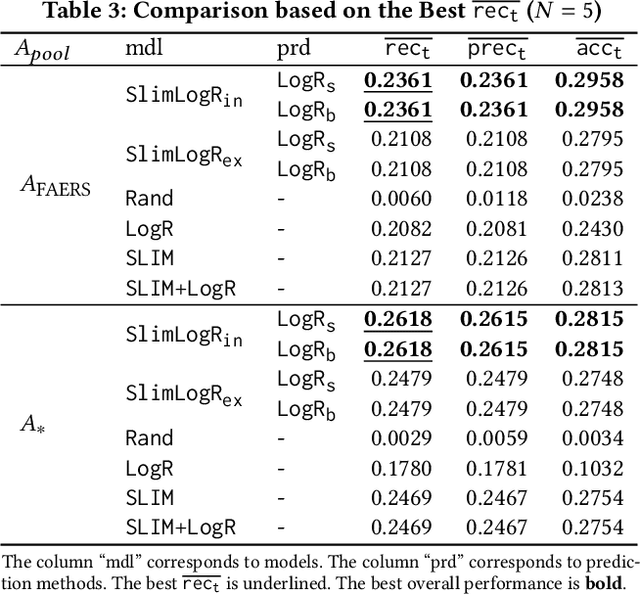
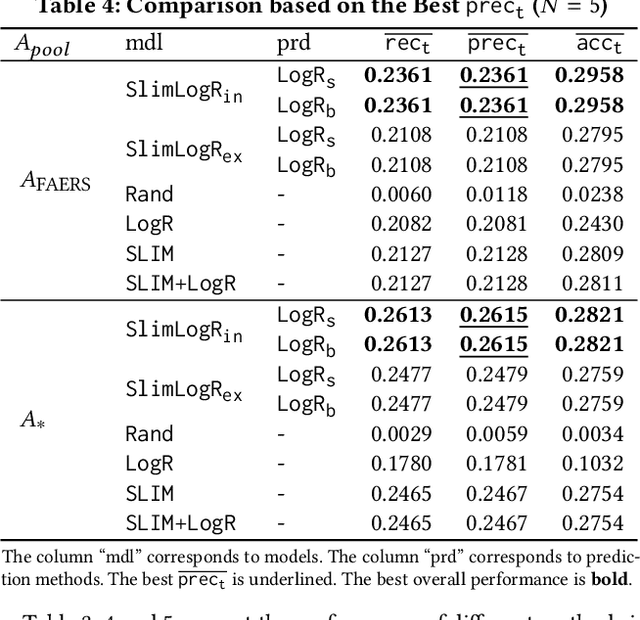
Abstract:Adverse drug reactions (ADRs) induced from high-order drug-drug interactions (DDIs) due to polypharmacy represent a significant public health problem. In this paper, we formally formulate the to-avoid and safe (with respect to ADRs) drug recommendation problems when multiple drugs have been taken simultaneously. We develop a joint model with a recommendation component and an ADR label prediction component to recommend for a prescription a set of to-avoid drugs that will induce ADRs if taken together with the prescription. We also develop real drug-drug interaction datasets and corresponding evaluation protocols. Our experimental results on real datasets demonstrate the strong performance of the joint model compared to other baseline methods.
Monitoring Potential Drug Interactions and Reactions via Network Analysis of Instagram User Timelines
Jan 14, 2016
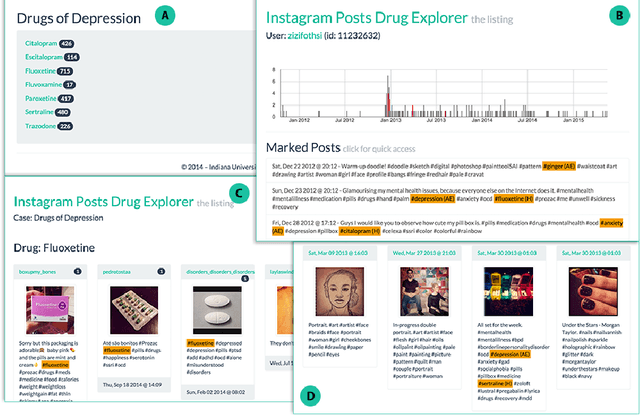
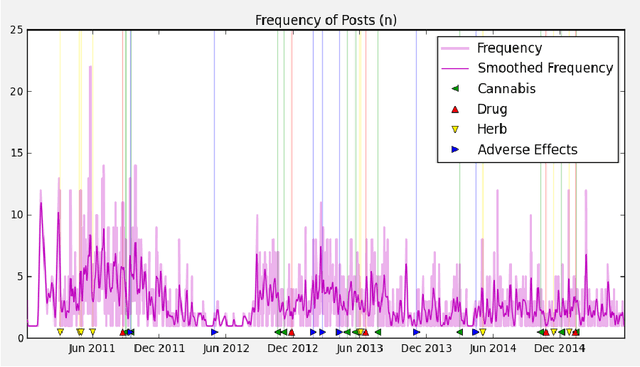
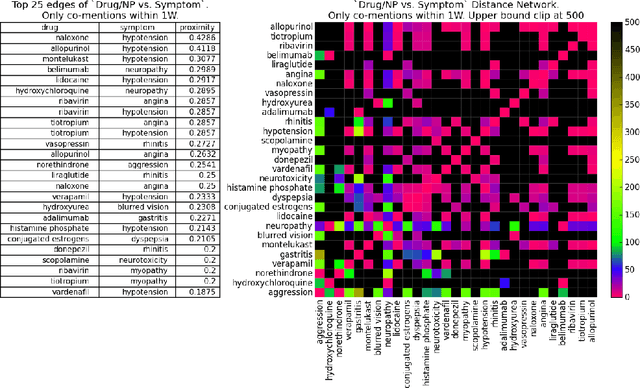
Abstract:Much recent research aims to identify evidence for Drug-Drug Interactions (DDI) and Adverse Drug reactions (ADR) from the biomedical scientific literature. In addition to this "Bibliome", the universe of social media provides a very promising source of large-scale data that can help identify DDI and ADR in ways that have not been hitherto possible. Given the large number of users, analysis of social media data may be useful to identify under-reported, population-level pathology associated with DDI, thus further contributing to improvements in population health. Moreover, tapping into this data allows us to infer drug interactions with natural products--including cannabis--which constitute an array of DDI very poorly explored by biomedical research thus far. Our goal is to determine the potential of Instagram for public health monitoring and surveillance for DDI, ADR, and behavioral pathology at large. Using drug, symptom, and natural product dictionaries for identification of the various types of DDI and ADR evidence, we have collected ~7000 timelines. We report on 1) the development of a monitoring tool to easily observe user-level timelines associated with drug and symptom terms of interest, and 2) population-level behavior via the analysis of co-occurrence networks computed from user timelines at three different scales: monthly, weekly, and daily occurrences. Analysis of these networks further reveals 3) drug and symptom direct and indirect associations with greater support in user timelines, as well as 4) clusters of symptoms and drugs revealed by the collective behavior of the observed population. This demonstrates that Instagram contains much drug- and pathology specific data for public health monitoring of DDI and ADR, and that complex network analysis provides an important toolbox to extract health-related associations and their support from large-scale social media data.
Extraction of Pharmacokinetic Evidence of Drug-drug Interactions from the Literature
May 18, 2015Abstract:Drug-drug interaction (DDI) is a major cause of morbidity and mortality and a subject of intense scientific interest. Biomedical literature mining can aid DDI research by extracting evidence for large numbers of potential interactions from published literature and clinical databases. Though DDI is investigated in domains ranging in scale from intracellular biochemistry to human populations, literature mining has not been used to extract specific types of experimental evidence, which are reported differently for distinct experimental goals. We focus on pharmacokinetic evidence for DDI, essential for identifying causal mechanisms of putative interactions and as input for further pharmacological and pharmaco-epidemiology investigations. We used manually curated corpora of PubMed abstracts and annotated sentences to evaluate the efficacy of literature mining on two tasks: first, identifying PubMed abstracts containing pharmacokinetic evidence of DDIs; second, extracting sentences containing such evidence from abstracts. We implemented a text mining pipeline and evaluated it using several linear classifiers and a variety of feature transforms. The most important textual features in the abstract and sentence classification tasks were analyzed. We also investigated the performance benefits of using features derived from PubMed metadata fields, various publicly available named entity recognizers, and pharmacokinetic dictionaries. Several classifiers performed very well in distinguishing relevant and irrelevant abstracts (reaching F1~=0.93, MCC~=0.74, iAUC~=0.99) and sentences (F1~=0.76, MCC~=0.65, iAUC~=0.83). We found that word bigram features were important for achieving optimal classifier performance and that features derived from Medical Subject Headings (MeSH) terms significantly improved abstract classification. ...
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge