Seungyeon Lee
A Deep Subgrouping Framework for Precision Drug Repurposing via Emulating Clinical Trials on Real-world Patient Data
Dec 29, 2024Abstract:Drug repurposing identifies new therapeutic uses for existing drugs, reducing the time and costs compared to traditional de novo drug discovery. Most existing drug repurposing studies using real-world patient data often treat the entire population as homogeneous, ignoring the heterogeneity of treatment responses across patient subgroups. This approach may overlook promising drugs that benefit specific subgroups but lack notable treatment effects across the entire population, potentially limiting the number of repurposable candidates identified. To address this, we introduce STEDR, a novel drug repurposing framework that integrates subgroup analysis with treatment effect estimation. Our approach first identifies repurposing candidates by emulating multiple clinical trials on real-world patient data and then characterizes patient subgroups by learning subgroup-specific treatment effects. We deploy \model to Alzheimer's Disease (AD), a condition with few approved drugs and known heterogeneity in treatment responses. We emulate trials for over one thousand medications on a large-scale real-world database covering over 8 million patients, identifying 14 drug candidates with beneficial effects to AD in characterized subgroups. Experiments demonstrate STEDR's superior capability in identifying repurposing candidates compared to existing approaches. Additionally, our method can characterize clinically relevant patient subgroups associated with important AD-related risk factors, paving the way for precision drug repurposing.
Network Representation Learning for Biophysical Neural Network Analysis
Oct 15, 2024
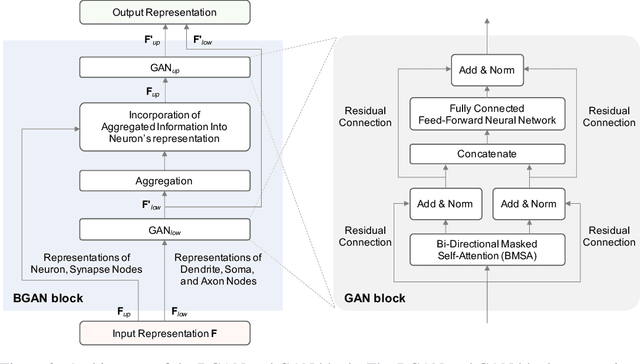
Abstract:The analysis of biophysical neural networks (BNNs) has been a longstanding focus in computational neuroscience. A central yet unresolved challenge in BNN analysis lies in deciphering the correlations between neuronal and synaptic dynamics, their connectivity patterns, and learning process. To address this, we introduce a novel BNN analysis framework grounded in network representation learning (NRL), which leverages attention scores to uncover intricate correlations between network components and their features. Our framework integrates a new computational graph (CG)-based BNN representation, a bio-inspired graph attention network (BGAN) that enables multiscale correlation analysis across BNN representations, and an extensive BNN dataset. The CG-based representation captures key computational features, information flow, and structural relationships underlying neuronal and synaptic dynamics, while BGAN reflects the compositional structure of neurons, including dendrites, somas, and axons, as well as bidirectional information flows between BNN components. The dataset comprises publicly available models from ModelDB, reconstructed using the Python and standardized in NeuroML format, and is augmented with data derived from canonical neuron and synapse models. To our knowledge, this study is the first to apply an NRL-based approach to the full spectrum of BNNs and their analysis.
Heterogeneous treatment effect estimation with subpopulation identification for personalized medicine in opioid use disorder
Jan 30, 2024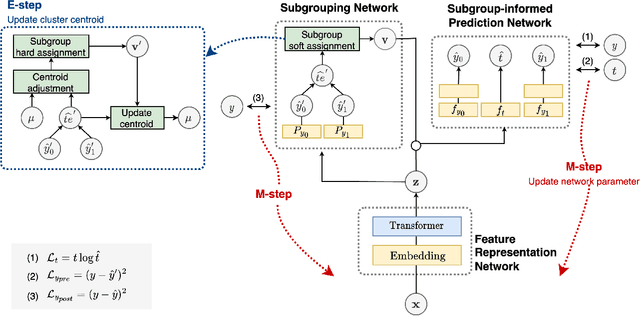
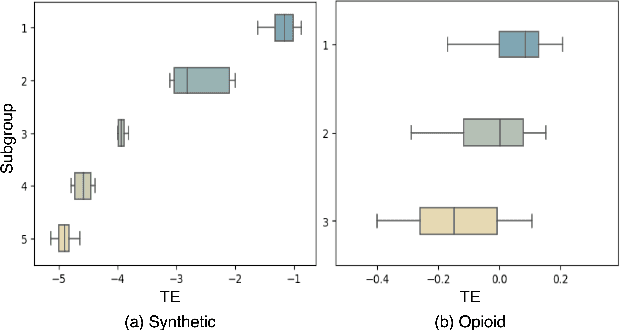
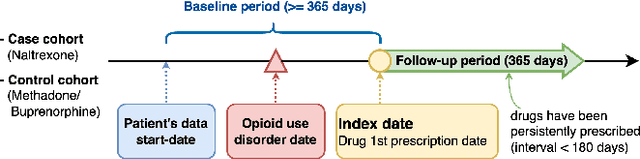
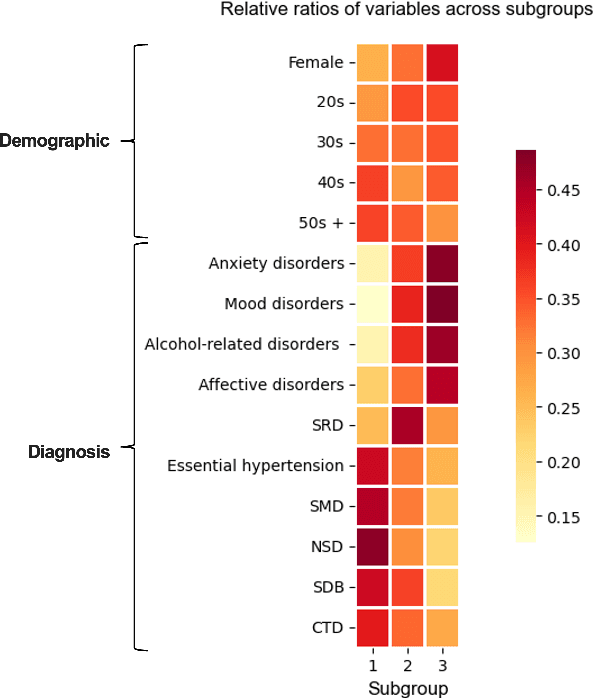
Abstract:Deep learning models have demonstrated promising results in estimating treatment effects (TEE). However, most of them overlook the variations in treatment outcomes among subgroups with distinct characteristics. This limitation hinders their ability to provide accurate estimations and treatment recommendations for specific subgroups. In this study, we introduce a novel neural network-based framework, named SubgroupTE, which incorporates subgroup identification and treatment effect estimation. SubgroupTE identifies diverse subgroups and simultaneously estimates treatment effects for each subgroup, improving the treatment effect estimation by considering the heterogeneity of treatment responses. Comparative experiments on synthetic data show that SubgroupTE outperforms existing models in treatment effect estimation. Furthermore, experiments on a real-world dataset related to opioid use disorder (OUD) demonstrate the potential of our approach to enhance personalized treatment recommendations for OUD patients.
SubgroupTE: Advancing Treatment Effect Estimation with Subgroup Identification
Jan 22, 2024
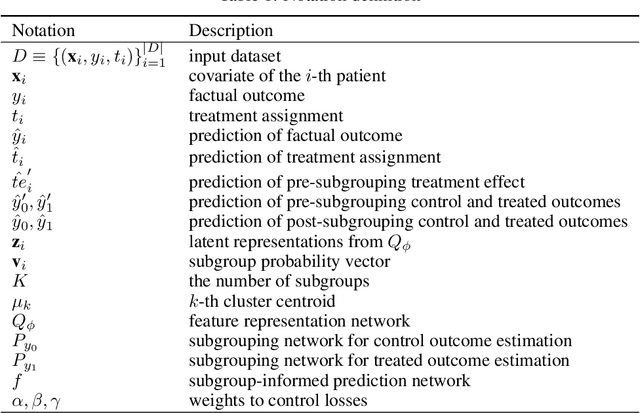
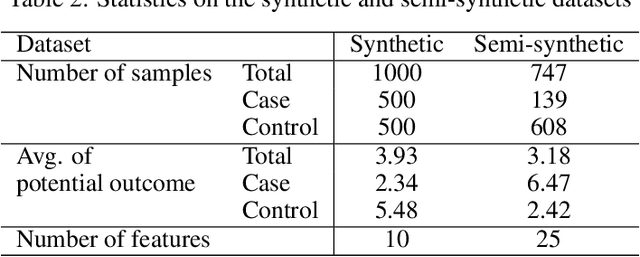
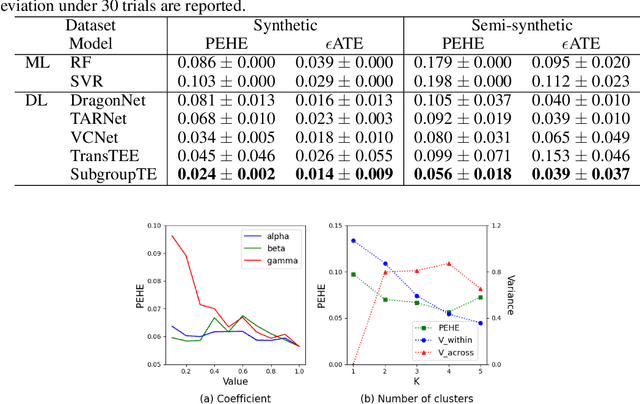
Abstract:Precise estimation of treatment effects is crucial for evaluating intervention effectiveness. While deep learning models have exhibited promising performance in learning counterfactual representations for treatment effect estimation (TEE), a major limitation in most of these models is that they treat the entire population as a homogeneous group, overlooking the diversity of treatment effects across potential subgroups that have varying treatment effects. This limitation restricts the ability to precisely estimate treatment effects and provide subgroup-specific treatment recommendations. In this paper, we propose a novel treatment effect estimation model, named SubgroupTE, which incorporates subgroup identification in TEE. SubgroupTE identifies heterogeneous subgroups with different treatment responses and more precisely estimates treatment effects by considering subgroup-specific causal effects. In addition, SubgroupTE iteratively optimizes subgrouping and treatment effect estimation networks to enhance both estimation and subgroup identification. Comprehensive experiments on the synthetic and semi-synthetic datasets exhibit the outstanding performance of SubgroupTE compared with the state-of-the-art models on treatment effect estimation. Additionally, a real-world study demonstrates the capabilities of SubgroupTE in enhancing personalized treatment recommendations for patients with opioid use disorder (OUD) by advancing treatment effect estimation with subgroup identification.
Domain Invariant Representation Learning and Sleep Dynamics Modeling for Automatic Sleep Staging
Dec 09, 2023Abstract:Sleep staging has become a critical task in diagnosing and treating sleep disorders to prevent sleep related diseases. With growing large scale sleep databases, significant progress has been made toward automatic sleep staging. However, previous studies face critical problems in sleep studies; the heterogeneity of subjects' physiological signals, the inability to extract meaningful information from unlabeled data to improve predictive performances, the difficulty in modeling correlations between sleep stages, and the lack of an effective mechanism to quantify predictive uncertainty. In this study, we propose a neural network based sleep staging model, DREAM, to learn domain generalized representations from physiological signals and models sleep dynamics. DREAM learns sleep related and subject invariant representations from diverse subjects' sleep signals and models sleep dynamics by capturing interactions between sequential signal segments and between sleep stages. We conducted a comprehensive empirical study to demonstrate the superiority of DREAM, including sleep stage prediction experiments, a case study, the usage of unlabeled data, and uncertainty. Notably, the case study validates DREAM's ability to learn generalized decision function for new subjects, especially in case there are differences between testing and training subjects. Uncertainty quantification shows that DREAM provides prediction uncertainty, making the model reliable and helping sleep experts in real world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge