Luis M. Rocha
Refinement of an Epilepsy Dictionary through Human Annotation of Health-related posts on Instagram
May 14, 2024Abstract:We used a dictionary built from biomedical terminology extracted from various sources such as DrugBank, MedDRA, MedlinePlus, TCMGeneDIT, to tag more than 8 million Instagram posts by users who have mentioned an epilepsy-relevant drug at least once, between 2010 and early 2016. A random sample of 1,771 posts with 2,947 term matches was evaluated by human annotators to identify false-positives. OpenAI's GPT series models were compared against human annotation. Frequent terms with a high false-positive rate were removed from the dictionary. Analysis of the estimated false-positive rates of the annotated terms revealed 8 ambiguous terms (plus synonyms) used in Instagram posts, which were removed from the original dictionary. To study the effect of removing those terms, we constructed knowledge networks using the refined and the original dictionaries and performed an eigenvector-centrality analysis on both networks. We show that the refined dictionary thus produced leads to a significantly different rank of important terms, as measured by their eigenvector-centrality of the knowledge networks. Furthermore, the most important terms obtained after refinement are of greater medical relevance. In addition, we show that OpenAI's GPT series models fare worse than human annotators in this task.
myAURA: Personalized health library for epilepsy management via knowledge graph sparsification and visualization
May 08, 2024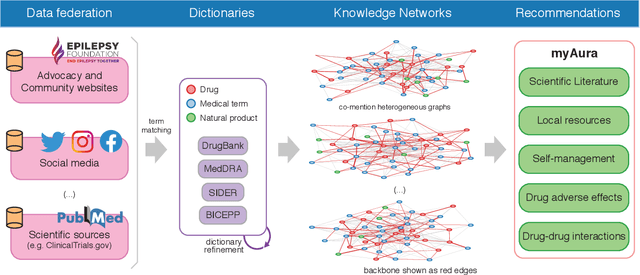
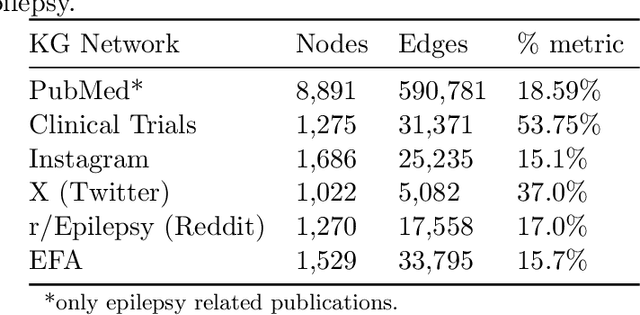
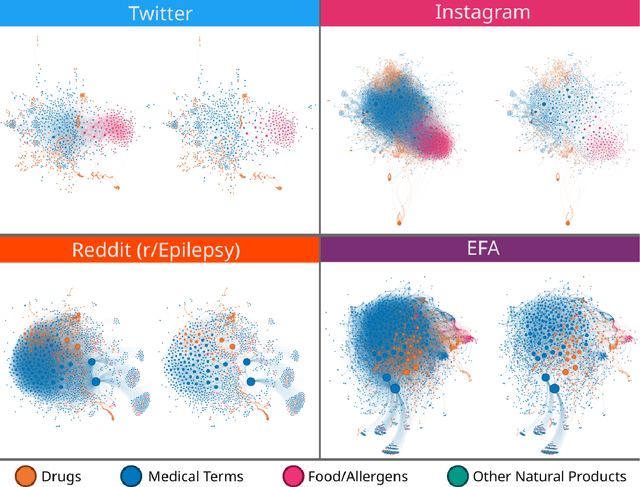
Abstract:Objective: We report the development of the patient-centered myAURA application and suite of methods designed to aid epilepsy patients, caregivers, and researchers in making decisions about care and self-management. Materials and Methods: myAURA rests on the federation of an unprecedented collection of heterogeneous data resources relevant to epilepsy, such as biomedical databases, social media, and electronic health records. A generalizable, open-source methodology was developed to compute a multi-layer knowledge graph linking all this heterogeneous data via the terms of a human-centered biomedical dictionary. Results: The power of the approach is first exemplified in the study of the drug-drug interaction phenomenon. Furthermore, we employ a novel network sparsification methodology using the metric backbone of weighted graphs, which reveals the most important edges for inference, recommendation, and visualization, such as pharmacology factors patients discuss on social media. The network sparsification approach also allows us to extract focused digital cohorts from social media whose discourse is more relevant to epilepsy or other biomedical problems. Finally, we present our patient-centered design and pilot-testing of myAURA, including its user interface, based on focus groups and other stakeholder input. Discussion: The ability to search and explore myAURA's heterogeneous data sources via a sparsified multi-layer knowledge graph, as well as the combination of those layers in a single map, are useful features for integrating relevant information for epilepsy. Conclusion: Our stakeholder-driven, scalable approach to integrate traditional and non-traditional data sources, enables biomedical discovery and data-powered patient self-management in epilepsy, and is generalizable to other chronic conditions.
The distance backbone of complex networks
Mar 08, 2021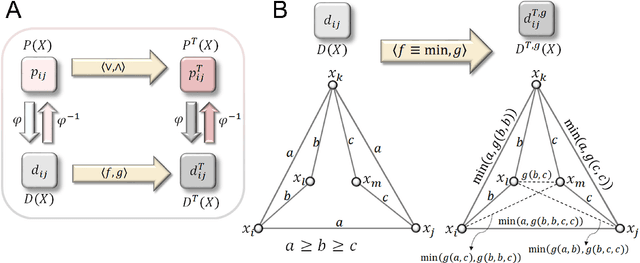
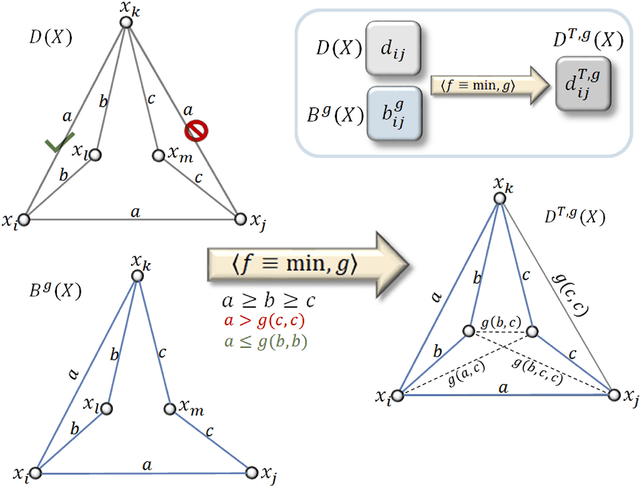

Abstract:Redundancy needs more precise characterization as it is a major factor in the evolution and robustness of networks of multivariate interactions. We investigate the complexity of such interactions by inferring a connection transitivity that includes all possible measures of path length for weighted graphs. The result, without breaking the graph into smaller components, is a distance backbone subgraph sufficient to compute all shortest paths. This is important for understanding the dynamics of spread and communication phenomena in real-world networks. The general methodology we formally derive yields a principled graph reduction technique and provides a finer characterization of the triangular geometry of shortest paths. We demonstrate that the distance backbone is very small in large networks across domains ranging from air traffic to the human brain connectome, revealing that network robustness to attacks and failures seems to stem from surprisingly vast amounts of redundancy.
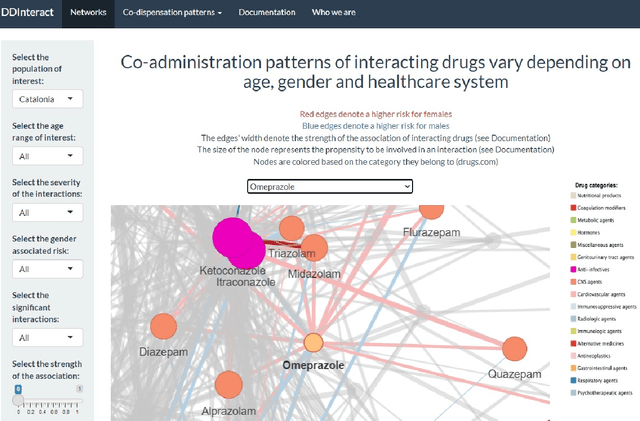
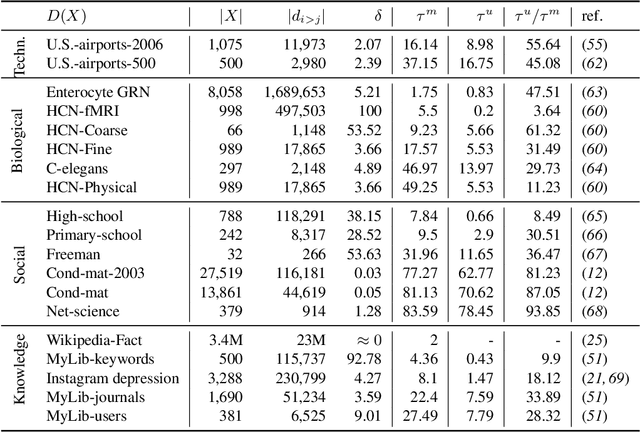
City-wide Analysis of Electronic Health Records Reveals Gender and Age Biases in the Administration of Known Drug-Drug Interactions
Mar 09, 2018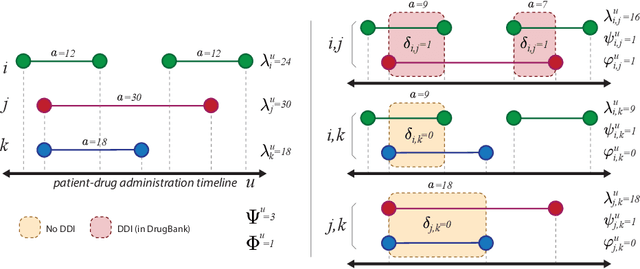



Abstract:From a public-health perspective, the occurrence of drug-drug-interactions (DDI) from multiple drug prescriptions is a serious problem, especially in the elderly population. This is true both for individuals and the system itself since patients with complications due to DDI will likely re-enter the system at a costlier level. We conducted an 18-month study of DDI occurrence in Blumenau (Brazil; pop. 340,000) using city-wide drug dispensing data from both primary and secondary-care level. Our goal is also to identify possible risk factors in a large population, ultimately characterizing the burden of DDI for patients, doctors and the public system itself. We found 181 distinct DDI being prescribed concomitantly to almost 5% of the city population. We also discovered that women are at a 60% risk increase of DDI when compared to men, while only having a 6% co-administration risk increase. Analysis of the DDI co-occurrence network reveals which DDI pairs are most associated with the observed greater DDI risk for females, demonstrating that contraception and hormone therapy are not the main culprits of the gender disparity, which is maximized after the reproductive years. Furthermore, DDI risk increases dramatically with age, with patients age 70-79 having a 50-fold risk increase in comparison to patients aged 0-19. Interestingly, several null models demonstrate that this risk increase is not due to increased polypharmacy with age. Finally, we demonstrate that while the number of drugs and co-administrations help predict a patient's number of DDI ($R^2=.413$), they are not sufficient to flag these patients accurately, which we achieve by training classifiers with additional data (MCC=.83,F1=.72). These results demonstrate that accurate warning systems for known DDI can be devised for public and private systems alike, resulting in substantial prevention of DDI-related ADR and savings.
Monitoring Potential Drug Interactions and Reactions via Network Analysis of Instagram User Timelines
Jan 14, 2016
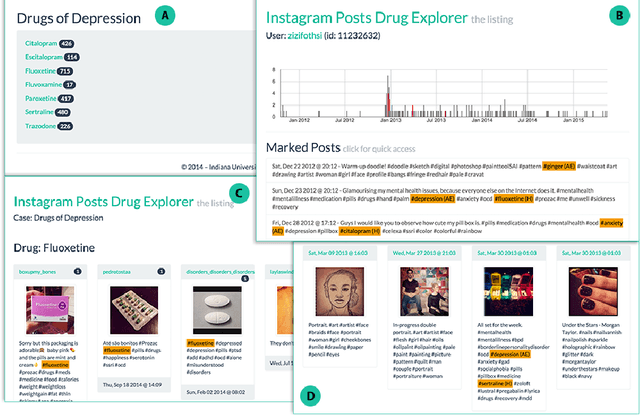
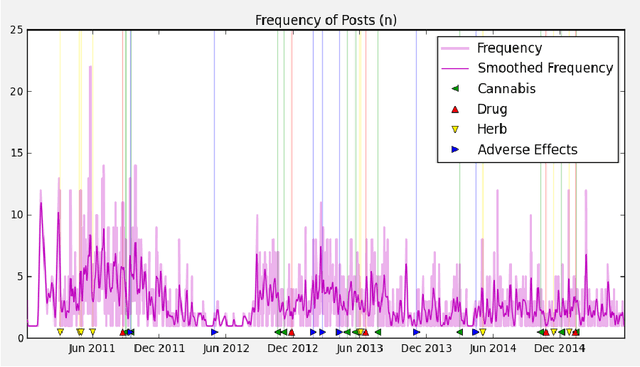
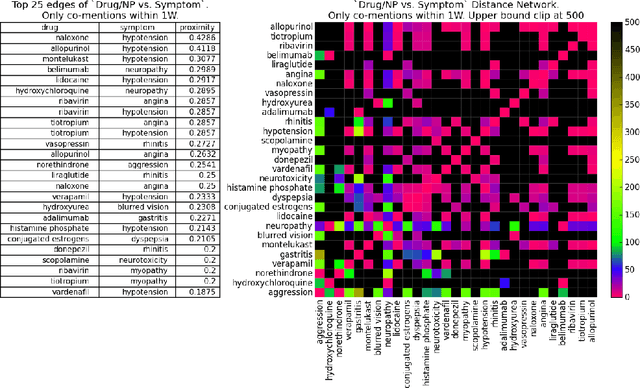
Abstract:Much recent research aims to identify evidence for Drug-Drug Interactions (DDI) and Adverse Drug reactions (ADR) from the biomedical scientific literature. In addition to this "Bibliome", the universe of social media provides a very promising source of large-scale data that can help identify DDI and ADR in ways that have not been hitherto possible. Given the large number of users, analysis of social media data may be useful to identify under-reported, population-level pathology associated with DDI, thus further contributing to improvements in population health. Moreover, tapping into this data allows us to infer drug interactions with natural products--including cannabis--which constitute an array of DDI very poorly explored by biomedical research thus far. Our goal is to determine the potential of Instagram for public health monitoring and surveillance for DDI, ADR, and behavioral pathology at large. Using drug, symptom, and natural product dictionaries for identification of the various types of DDI and ADR evidence, we have collected ~7000 timelines. We report on 1) the development of a monitoring tool to easily observe user-level timelines associated with drug and symptom terms of interest, and 2) population-level behavior via the analysis of co-occurrence networks computed from user timelines at three different scales: monthly, weekly, and daily occurrences. Analysis of these networks further reveals 3) drug and symptom direct and indirect associations with greater support in user timelines, as well as 4) clusters of symptoms and drugs revealed by the collective behavior of the observed population. This demonstrates that Instagram contains much drug- and pathology specific data for public health monitoring of DDI and ADR, and that complex network analysis provides an important toolbox to extract health-related associations and their support from large-scale social media data.
Extraction of Pharmacokinetic Evidence of Drug-drug Interactions from the Literature
May 18, 2015Abstract:Drug-drug interaction (DDI) is a major cause of morbidity and mortality and a subject of intense scientific interest. Biomedical literature mining can aid DDI research by extracting evidence for large numbers of potential interactions from published literature and clinical databases. Though DDI is investigated in domains ranging in scale from intracellular biochemistry to human populations, literature mining has not been used to extract specific types of experimental evidence, which are reported differently for distinct experimental goals. We focus on pharmacokinetic evidence for DDI, essential for identifying causal mechanisms of putative interactions and as input for further pharmacological and pharmaco-epidemiology investigations. We used manually curated corpora of PubMed abstracts and annotated sentences to evaluate the efficacy of literature mining on two tasks: first, identifying PubMed abstracts containing pharmacokinetic evidence of DDIs; second, extracting sentences containing such evidence from abstracts. We implemented a text mining pipeline and evaluated it using several linear classifiers and a variety of feature transforms. The most important textual features in the abstract and sentence classification tasks were analyzed. We also investigated the performance benefits of using features derived from PubMed metadata fields, various publicly available named entity recognizers, and pharmacokinetic dictionaries. Several classifiers performed very well in distinguishing relevant and irrelevant abstracts (reaching F1~=0.93, MCC~=0.74, iAUC~=0.99) and sentences (F1~=0.76, MCC~=0.65, iAUC~=0.83). We found that word bigram features were important for achieving optimal classifier performance and that features derived from Medical Subject Headings (MeSH) terms significantly improved abstract classification. ...
Prediction and Modularity in Dynamical Systems
Jan 16, 2015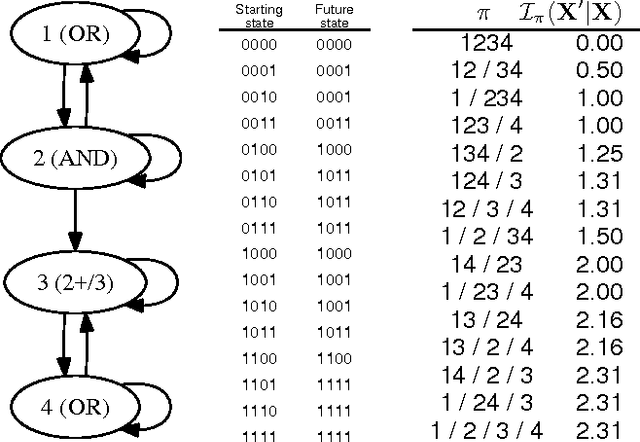
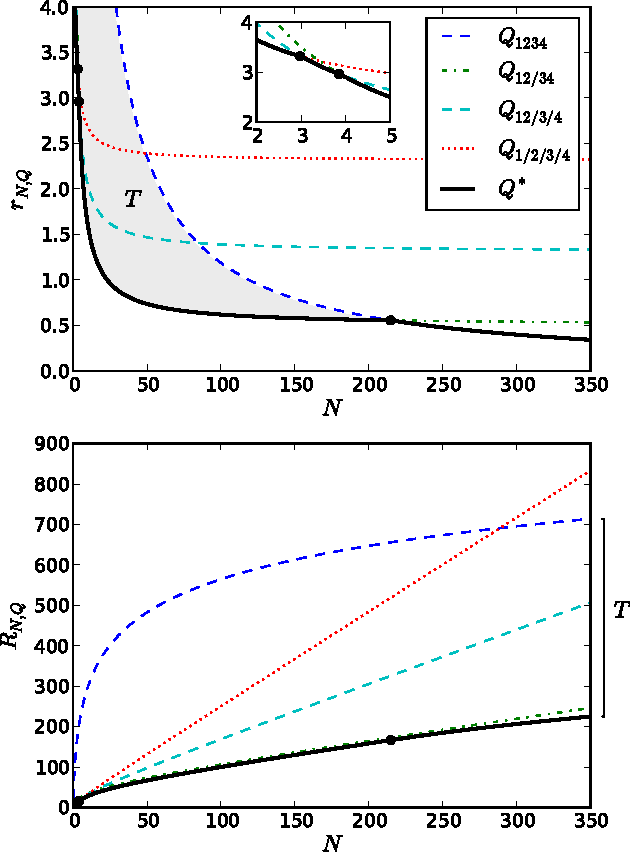
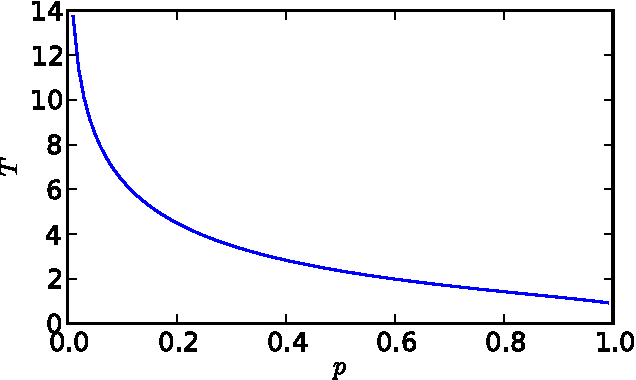
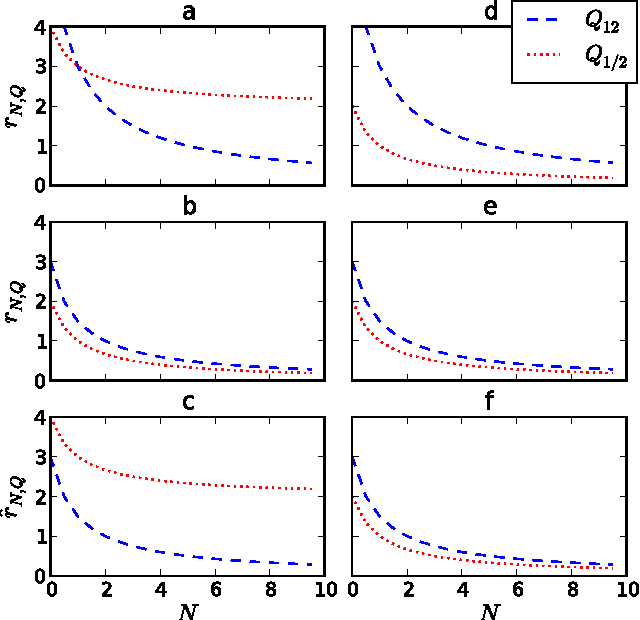
Abstract:Identifying and understanding modular organizations is centrally important in the study of complex systems. Several approaches to this problem have been advanced, many framed in information-theoretic terms. Our treatment starts from the complementary point of view of statistical modeling and prediction of dynamical systems. It is known that for finite amounts of training data, simpler models can have greater predictive power than more complex ones. We use the trade-off between model simplicity and predictive accuracy to generate optimal multiscale decompositions of dynamical networks into weakly-coupled, simple modules. State-dependent and causal versions of our method are also proposed.
Designing a minimalist socially aware robotic agent for the home
Jun 26, 2014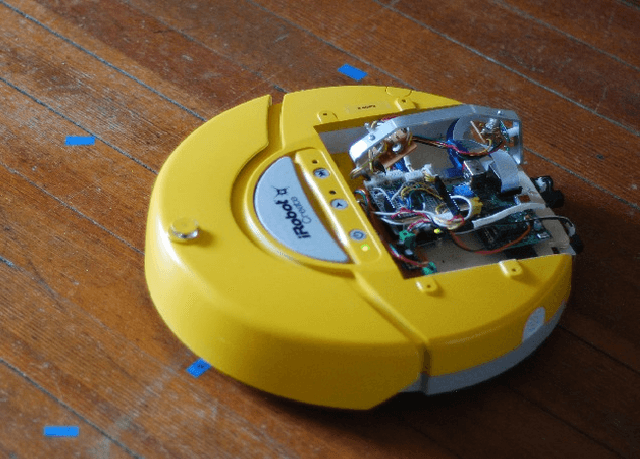
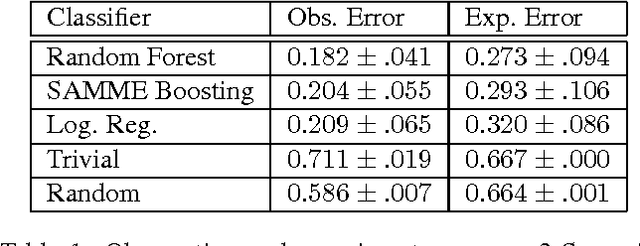
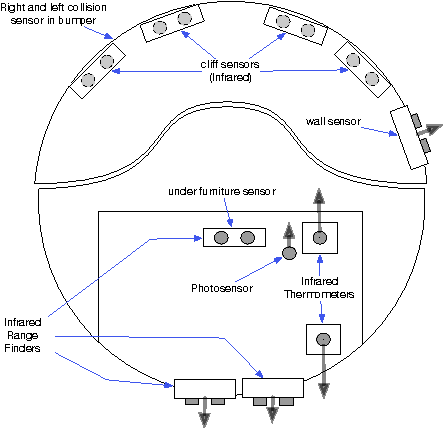
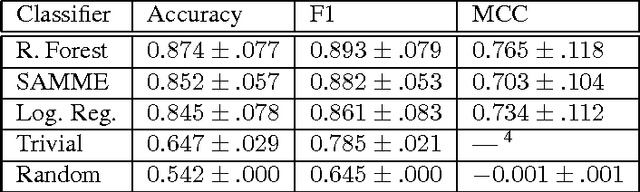
Abstract:We present a minimalist social robot that relies on long timeseries of low resolution data such as mechanical vibration, temperature, lighting, sounds and collisions. Our goal is to develop an experimental system for growing socially situated robotic agents whose behavioral repertoire is subsumed by the social order of the space. To get there we are designing robots that use their simple sensors and motion feedback routines to recognize different classes of human activity and then associate to each class a range of appropriate behaviors. We use the Katie Family of robots, built on the iRobot Create platform, an Arduino Uno, and a Raspberry Pi. We describe its sensor abilities and exploratory tests that allow us to develop hypotheses about what objects (sensor data) correspond to something known and observable by a human subject. We use machine learning methods to classify three social scenarios from over a hundred experiments, demonstrating that it is possible to detect social situations with high accuracy, using the low-resolution sensors from our minimalist robot.
Evaluation of linear classifiers on articles containing pharmacokinetic evidence of drug-drug interactions
Oct 02, 2012



Abstract:Background. Drug-drug interaction (DDI) is a major cause of morbidity and mortality. [...] Biomedical literature mining can aid DDI research by extracting relevant DDI signals from either the published literature or large clinical databases. However, though drug interaction is an ideal area for translational research, the inclusion of literature mining methodologies in DDI workflows is still very preliminary. One area that can benefit from literature mining is the automatic identification of a large number of potential DDIs, whose pharmacological mechanisms and clinical significance can then be studied via in vitro pharmacology and in populo pharmaco-epidemiology. Experiments. We implemented a set of classifiers for identifying published articles relevant to experimental pharmacokinetic DDI evidence. These documents are important for identifying causal mechanisms behind putative drug-drug interactions, an important step in the extraction of large numbers of potential DDIs. We evaluate performance of several linear classifiers on PubMed abstracts, under different feature transformation and dimensionality reduction methods. In addition, we investigate the performance benefits of including various publicly-available named entity recognition features, as well as a set of internally-developed pharmacokinetic dictionaries. Results. We found that several classifiers performed well in distinguishing relevant and irrelevant abstracts. We found that the combination of unigram and bigram textual features gave better performance than unigram features alone, and also that normalization transforms that adjusted for feature frequency and document length improved classification. For some classifiers, such as linear discriminant analysis (LDA), proper dimensionality reduction had a large impact on performance. Finally, the inclusion of NER features and dictionaries was found not to help classification.
* Pacific Symposium on Biocomputing, 2013
A Linear Classifier Based on Entity Recognition Tools and a Statistical Approach to Method Extraction in the Protein-Protein Interaction Literature
Apr 22, 2011

Abstract:We participated, in the Article Classification and the Interaction Method subtasks (ACT and IMT, respectively) of the Protein-Protein Interaction task of the BioCreative III Challenge. For the ACT, we pursued an extensive testing of available Named Entity Recognition and dictionary tools, and used the most promising ones to extend our Variable Trigonometric Threshold linear classifier. For the IMT, we experimented with a primarily statistical approach, as opposed to employing a deeper natural language processing strategy. Finally, we also studied the benefits of integrating the method extraction approach that we have used for the IMT into the ACT pipeline. For the ACT, our linear article classifier leads to a ranking and classification performance significantly higher than all the reported submissions. For the IMT, our results are comparable to those of other systems, which took very different approaches. For the ACT, we show that the use of named entity recognition tools leads to a substantial improvement in the ranking and classification of articles relevant to protein-protein interaction. Thus, we show that our substantially expanded linear classifier is a very competitive classifier in this domain. Moreover, this classifier produces interpretable surfaces that can be understood as "rules" for human understanding of the classification. In terms of the IMT task, in contrast to other participants, our approach focused on identifying sentences that are likely to bear evidence for the application of a PPI detection method, rather than on classifying a document as relevant to a method. As BioCreative III did not perform an evaluation of the evidence provided by the system, we have conducted a separate assessment; the evaluators agree that our tool is indeed effective in detecting relevant evidence for PPI detection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge