Hagit Shatkay
Domain-informed neural networks for interaction localization within astroparticle experiments
Dec 15, 2021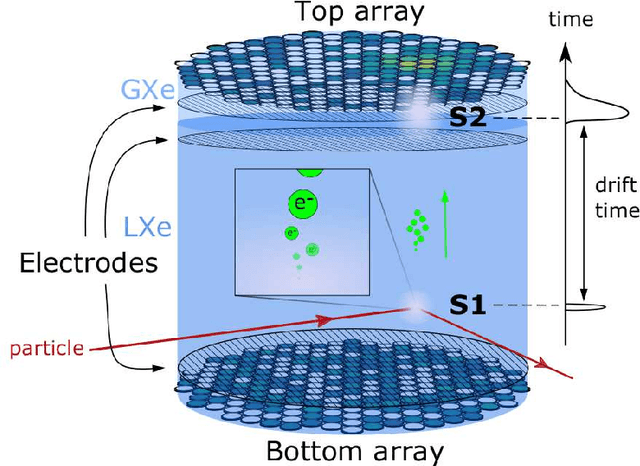
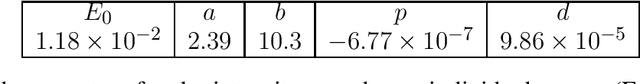
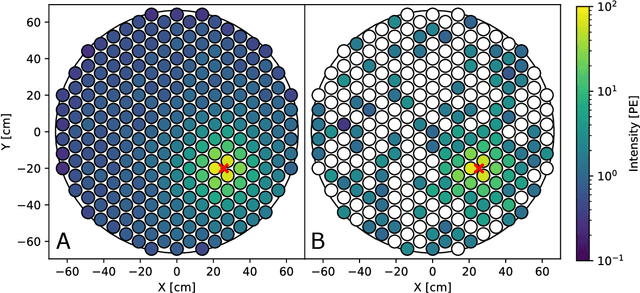
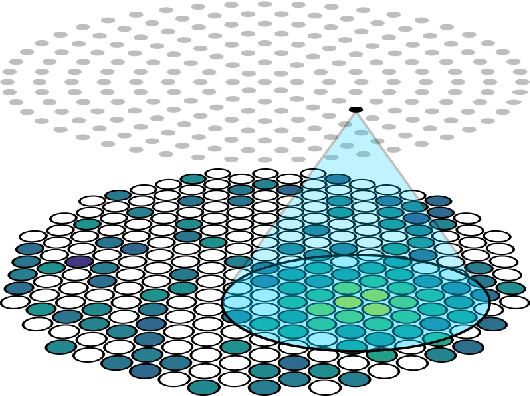
Abstract:This work proposes a domain-informed neural network architecture for experimental particle physics, using particle interaction localization with the time-projection chamber (TPC) technology for dark matter research as an example application. A key feature of the signals generated within the TPC is that they allow localization of particle interactions through a process called reconstruction. While multilayer perceptrons (MLPs) have emerged as a leading contender for reconstruction in TPCs, such a black-box approach does not reflect prior knowledge of the underlying scientific processes. This paper looks anew at neural network-based interaction localization and encodes prior detector knowledge, in terms of both signal characteristics and detector geometry, into the feature encoding and the output layers of a multilayer neural network. The resulting Domain-informed Neural Network (DiNN limits the receptive fields of the neurons in the initial feature encoding layers in order to account for the spatially localized nature of the signals produced within the TPC. This aspect of the DiNN, which has similarities with the emerging area of graph neural networks in that the neurons in the initial layers only connect to a handful of neurons in their succeeding layer, significantly reduces the number of parameters in the network in comparison to an MLP. In addition, in order to account for the detector geometry, the output layers of the network are modified using two geometric transformations to ensure the DiNN produces localizations within the interior of the detector. The end result is a neural network architecture that has 60% fewer parameters than an MLP, but that still achieves similar localization performance and provides a path to future architectural developments with improved performance because of their ability to encode additional domain knowledge into the architecture.
Identifying Ventricular Arrhythmias and Their Predictors by Applying Machine Learning Methods to Electronic Health Records in Patients With Hypertrophic Cardiomyopathy(HCM-VAr-Risk Model)
Sep 19, 2021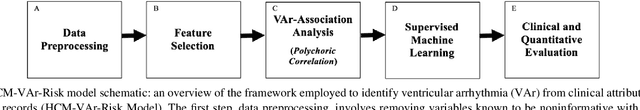
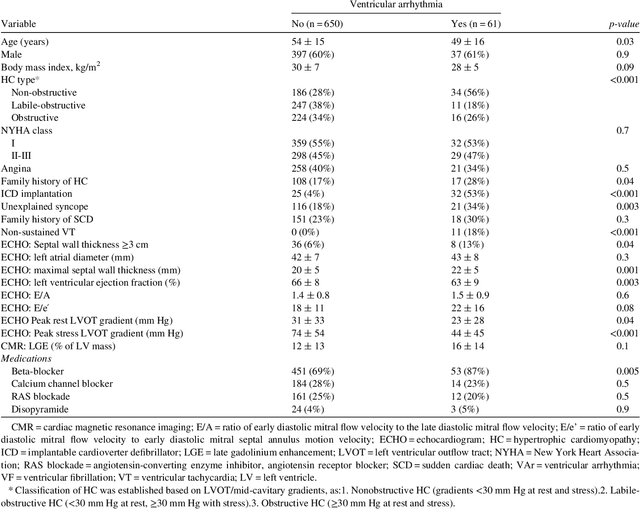
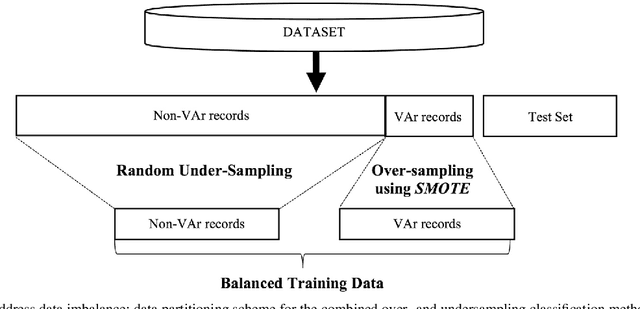
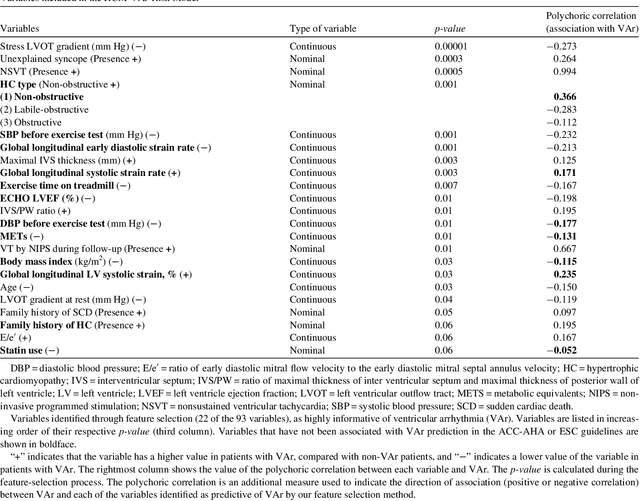
Abstract:Clinical risk stratification for sudden cardiac death (SCD) in hypertrophic cardiomyopathy (HC) employs rules derived from American College of Cardiology Foundation/American Heart Association (ACCF/AHA) guidelines or the HCM Risk-SCD model (C-index of 0.69), which utilize a few clinical variables. We assessed whether data-driven machine learning methods that consider a wider range of variables can effectively identify HC patients with ventricular arrhythmias (VAr) that lead to SCD. We scanned the electronic health records of 711 HC patients for sustained ventricular tachycardia or ventricular fibrillation. Patients with ventricular tachycardia or ventricular fibrillation (n = 61) were tagged as VAr cases and the remaining (n = 650) as non-VAr. The 2-sample t test and information gain criterion were used to identify the most informative clinical variables that distinguish VAr from non-VAr; patient records were reduced to include only these variables. Data imbalance stemming from low number of VAr cases was addressed by applying a combination of over- and under-sampling strategies.We trained and tested multiple classifiers under this sampling approach, showing effective classification. We evaluated 93 clinical variables, of which 22 proved predictive of VAr. The ensemble of logistic regression and naive Bayes classifiers, trained based on these 22 variables and corrected for data imbalance, was most effective in separating VAr from non-VAr cases (sensitivity = 0.73, specificity = 0.76, C-index = 0.83). Our method (HCM-VAr-Risk Model) identified 12 new predictors of VAr, in addition to 10 established SCD predictors. In conclusion, this is the first application of machine learning for identifying HC patients with VAr, using clinical attributes.
Machine Learning Methods for Identifying Atrial Fibrillation Cases and Their Predictors in Patients With Hypertrophic Cardiomyopathy: The HCM-AF-Risk Model
Sep 19, 2021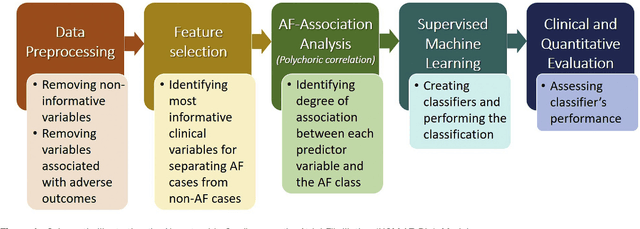
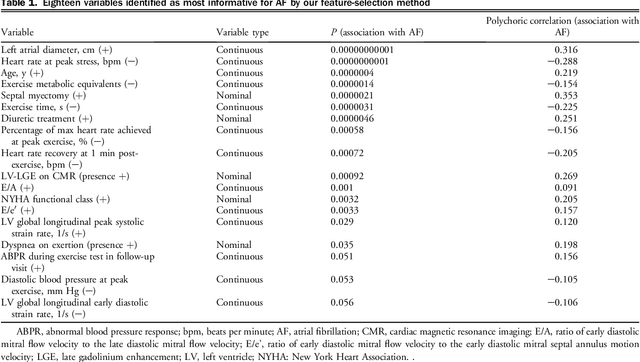
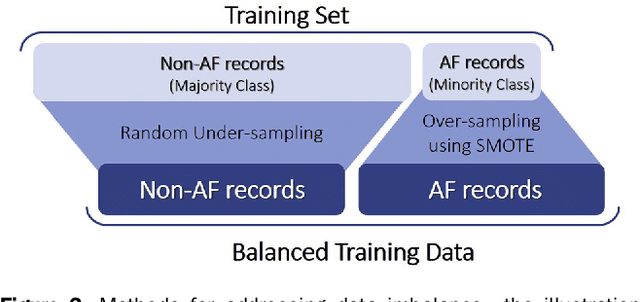
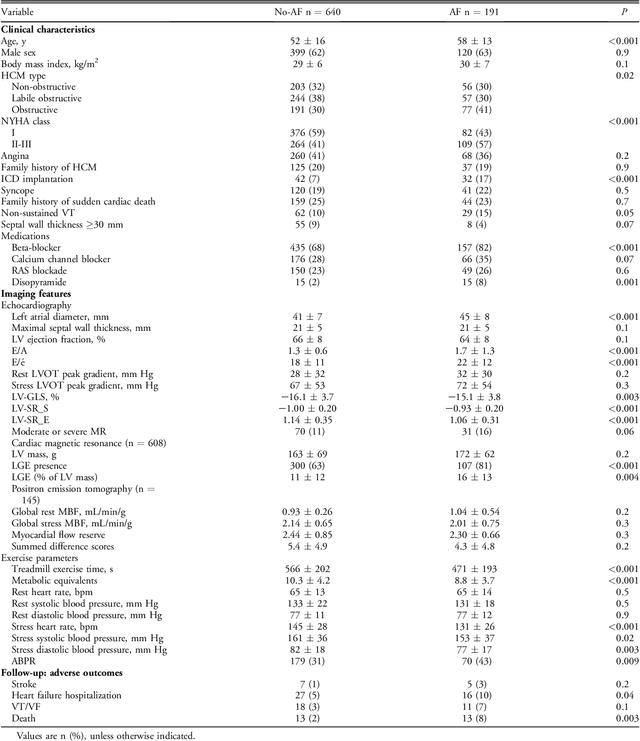
Abstract:Hypertrophic cardiomyopathy (HCM) patients have a high incidence of atrial fibrillation (AF) and increased stroke risk, even with low risk of congestive heart failure, hypertension, age, diabetes, previous stroke/transient ischemic attack scores. Hence, there is a need to understand the pathophysiology of AF and stroke in HCM. In this retrospective study, we develop and apply a data-driven, machine learning based method to identify AF cases, and clinical and imaging features associated with AF, using electronic health record data. HCM patients with documented paroxysmal/persistent/permanent AF (n = 191) were considered AF cases, and the remaining patients in sinus rhythm (n = 640) were tagged as No-AF. We evaluated 93 clinical variables and the most informative variables useful for distinguishing AF from No-AF cases were selected based on the 2-sample t test and the information gain criterion. We identified 18 highly informative variables that are positively (n = 11) and negatively (n = 7) correlated with AF in HCM. Next, patient records were represented via these 18 variables. Data imbalance resulting from the relatively low number of AF cases was addressed via a combination of oversampling and under-sampling strategies. We trained and tested multiple classifiers under this sampling approach, showing effective classification. Specifically, an ensemble of logistic regression and naive Bayes classifiers, trained based on the 18 variables and corrected for data imbalance, proved most effective for separating AF from No-AF cases (sensitivity = 0.74, specificity = 0.70, C-index = 0.80). Our model is the first machine learning based method for identification of AF cases in HCM. This model demonstrates good performance, addresses data imbalance, and suggests that AF is associated with a more severe cardiac HCM phenotype.
Co-occurrence of medical conditions: Exposing patterns through probabilistic topic modeling of SNOMED codes
Sep 19, 2021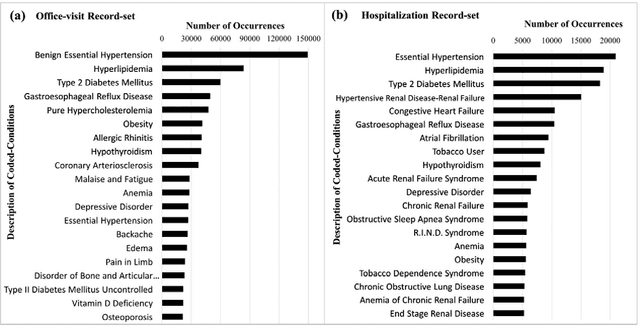
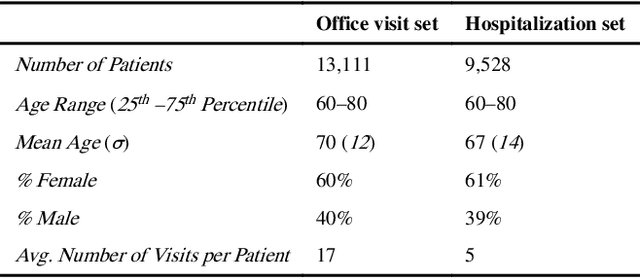
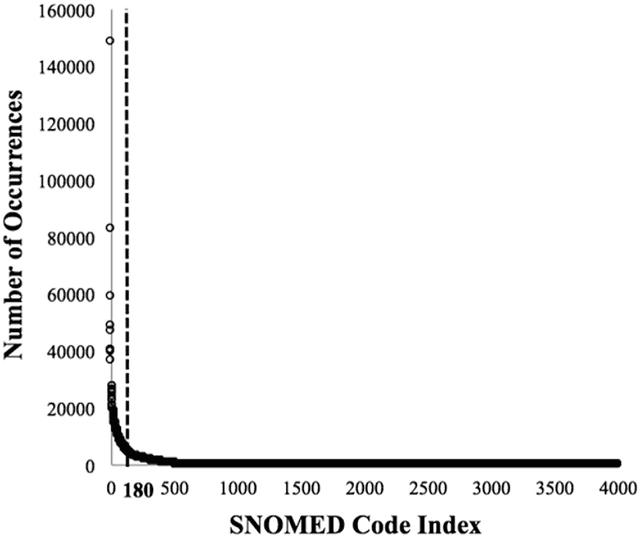
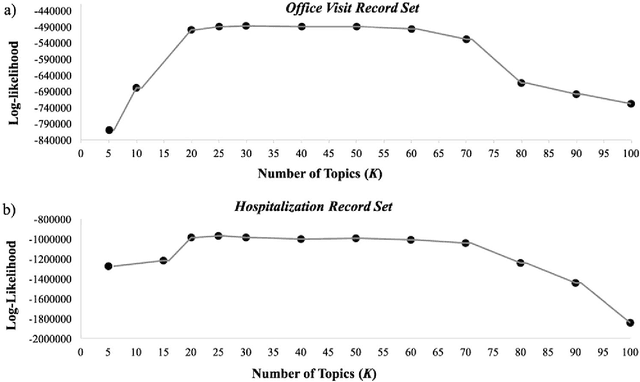
Abstract:Patients associated with multiple co-occurring health conditions often face aggravated complications and less favorable outcomes. Co-occurring conditions are especially prevalent among individuals suffering from kidney disease, an increasingly widespread condition affecting 13% of the general population in the US. This study aims to identify and characterize patterns of co-occurring medical conditions in patients employing a probabilistic framework. Specifically, we apply topic modeling in a non-traditional way to find associations across SNOMEDCT codes assigned and recorded in the EHRs of>13,000 patients diagnosed with kidney disease. Unlike most prior work on topic modeling, we apply the method to codes rather than to natural language. Moreover, we quantitatively evaluate the topics, assessing their tightness and distinctiveness, and also assess the medical validity of our results. Our experiments show that each topic is succinctly characterized by a few highly probable and unique disease codes, indicating that the topics are tight. Furthermore, inter-topic distance between each pair of topics is typically high, illustrating distinctiveness. Last, most coded conditions grouped together within a topic, are indeed reported to co-occur in the medical literature. Notably, our results uncover a few indirect associations among conditions that have hitherto not been reported as correlated in the medical literature.
Identifying Patterns of Associated-Conditions through Topic Models of Electronic Medical Records
Nov 17, 2017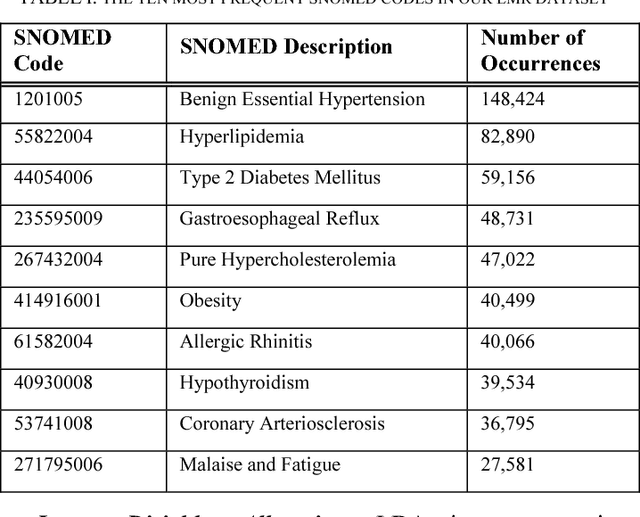
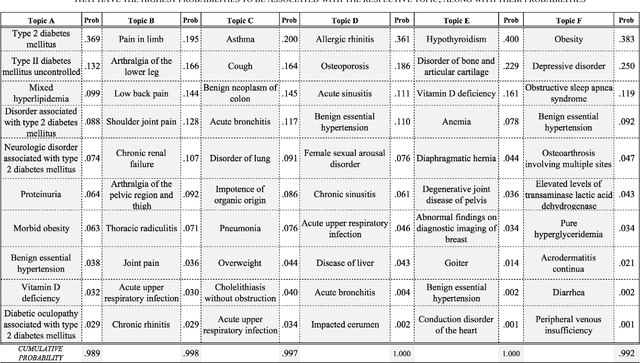
Abstract:Multiple adverse health conditions co-occurring in a patient are typically associated with poor prognosis and increased office or hospital visits. Developing methods to identify patterns of co-occurring conditions can assist in diagnosis. Thus identifying patterns of associations among co-occurring conditions is of growing interest. In this paper, we report preliminary results from a data-driven study, in which we apply a machine learning method, namely, topic modeling, to electronic medical records, aiming to identify patterns of associated conditions. Specifically, we use the well established latent dirichlet allocation, a method based on the idea that documents can be modeled as a mixture of latent topics, where each topic is a distribution over words. In our study, we adapt the LDA model to identify latent topics in patients' EMRs. We evaluate the performance of our method both qualitatively, and show that the obtained topics indeed align well with distinct medical phenomena characterized by co-occurring conditions.
Identifying Growth-Patterns in Children by Applying Cluster analysis to Electronic Medical Records
Aug 16, 2017

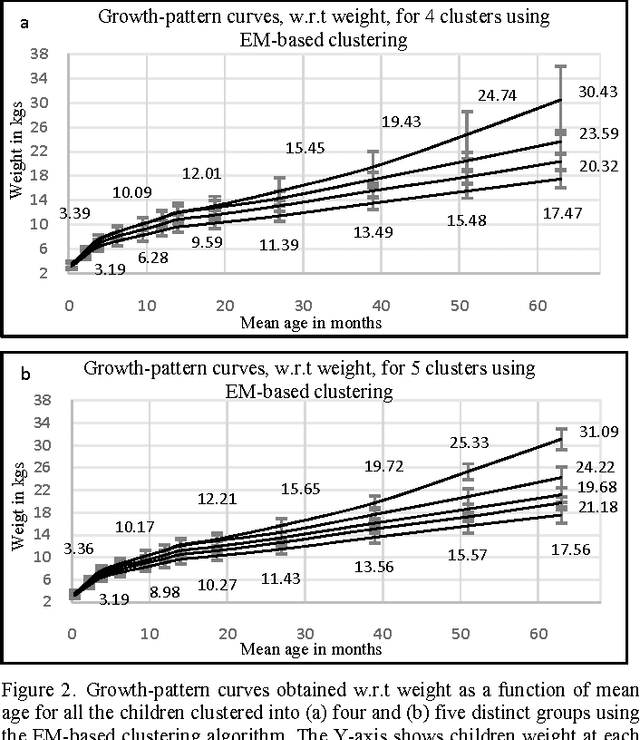

Abstract:Obesity is one of the leading health concerns in the United States. Researchers and health care providers are interested in understanding factors affecting obesity and detecting the likelihood of obesity as early as possible. In this paper, we set out to recognize children who have higher risk of obesity by identifying distinct growth patterns in them. This is done by using clustering methods, which group together children who share similar body measurements over a period of time. The measurements characterizing children within the same cluster are plotted as a function of age. We refer to these plots as growthpattern curves. We show that distinct growth-pattern curves are associated with different clusters and thus can be used to separate children into the topmost (heaviest), middle, or bottom-most cluster based on early growth measurements.
Protein (Multi-)Location Prediction: Using Location Inter-Dependencies in a Probabilistic Framework
Jul 30, 2013



Abstract:Knowing the location of a protein within the cell is important for understanding its function, role in biological processes, and potential use as a drug target. Much progress has been made in developing computational methods that predict single locations for proteins, assuming that proteins localize to a single location. However, it has been shown that proteins localize to multiple locations. While a few recent systems have attempted to predict multiple locations of proteins, they typically treat locations as independent or capture inter-dependencies by treating each locations-combination present in the training set as an individual location-class. We present a new method and a preliminary system we have developed that directly incorporates inter-dependencies among locations into the multiple-location-prediction process, using a collection of Bayesian network classifiers. We evaluate our system on a dataset of single- and multi-localized proteins. Our results, obtained by incorporating inter-dependencies are significantly higher than those obtained by classifiers that do not use inter-dependencies. The performance of our system on multi-localized proteins is comparable to a top performing system (YLoc+), without restricting predictions to be based only on location-combinations present in the training set.
Learning Hidden Markov Models with Geometrical Constraints
Jan 23, 2013



Abstract:Hidden Markov models (HMMs) and partially observable Markov decision processes (POMDPs) form a useful tool for modeling dynamical systems. They are particularly useful for representing environments such as road networks and office buildings, which are typical for robot navigation and planning. The work presented here is concerned with acquiring such models. We demonstrate how domain-specific information and constraints can be incorporated into the statistical estimation process, greatly improving the learned models in terms of the model quality, the number of iterations required for convergence and robustness to reduction in the amount of available data. We present new initialization heuristics which can be used even when the data suffers from cumulative rotational error, new update rules for the model parameters, as an instance of generalized EM, and a strategy for enforcing complete geometrical consistency in the model. Experimental results demonstrate the effectiveness of our approach for both simulated and real robot data, in traditionally hard-to-learn environments.
A Linear Classifier Based on Entity Recognition Tools and a Statistical Approach to Method Extraction in the Protein-Protein Interaction Literature
Apr 22, 2011

Abstract:We participated, in the Article Classification and the Interaction Method subtasks (ACT and IMT, respectively) of the Protein-Protein Interaction task of the BioCreative III Challenge. For the ACT, we pursued an extensive testing of available Named Entity Recognition and dictionary tools, and used the most promising ones to extend our Variable Trigonometric Threshold linear classifier. For the IMT, we experimented with a primarily statistical approach, as opposed to employing a deeper natural language processing strategy. Finally, we also studied the benefits of integrating the method extraction approach that we have used for the IMT into the ACT pipeline. For the ACT, our linear article classifier leads to a ranking and classification performance significantly higher than all the reported submissions. For the IMT, our results are comparable to those of other systems, which took very different approaches. For the ACT, we show that the use of named entity recognition tools leads to a substantial improvement in the ranking and classification of articles relevant to protein-protein interaction. Thus, we show that our substantially expanded linear classifier is a very competitive classifier in this domain. Moreover, this classifier produces interpretable surfaces that can be understood as "rules" for human understanding of the classification. In terms of the IMT task, in contrast to other participants, our approach focused on identifying sentences that are likely to bear evidence for the application of a PPI detection method, rather than on classifying a document as relevant to a method. As BioCreative III did not perform an evaluation of the evidence provided by the system, we have conducted a separate assessment; the evaluators agree that our tool is indeed effective in detecting relevant evidence for PPI detection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge