Michael Conover
Considerations for the Interpretation of Bias Measures of Word Embeddings
Jun 19, 2019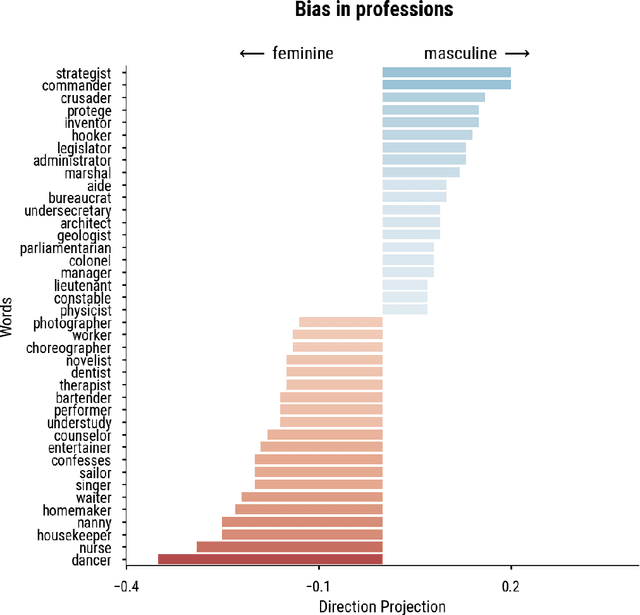
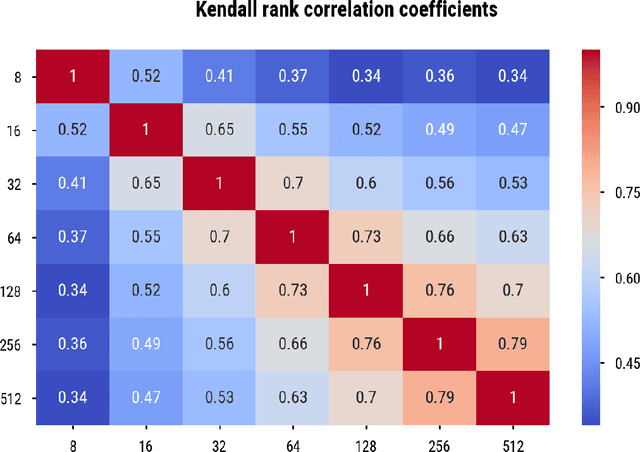
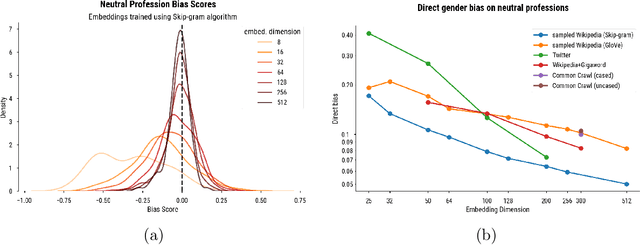
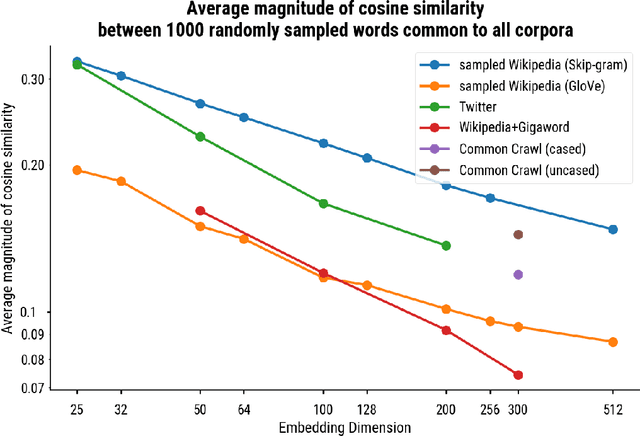
Abstract:Word embedding spaces are powerful tools for capturing latent semantic relationships between terms in corpora, and have become widely popular for building state-of-the-art natural language processing algorithms. However, studies have shown that societal biases present in text corpora may be incorporated into the word embedding spaces learned from them. Thus, there is an ethical concern that human-like biases contained in the corpora and their derived embedding spaces might be propagated, or even amplified with the usage of the biased embedding spaces in downstream applications. In an attempt to quantify these biases so that they may be better understood and studied, several bias metrics have been proposed. We explore the statistical properties of these proposed measures in the context of their cited applications as well as their supposed utilities. We find that there are caveats to the simple interpretation of these metrics as proposed. We find that the bias metric proposed by Bolukbasi et al. 2016 is highly sensitive to embedding hyper-parameter selection, and that in many cases, the variance due to the selection of some hyper-parameters is greater than the variance in the metric due to corpus selection, while in fewer cases the bias rankings of corpora vary with hyper-parameter selection. In light of these observations, it may be the case that bias estimates should not be thought to directly measure the properties of the underlying corpus, but rather the properties of the specific embedding spaces in question, particularly in the context of hyper-parameter selections used to generate them. Hence, bias metrics of spaces generated with differing hyper-parameters should be compared only with explicit consideration of the embedding-learning algorithms particular configurations.
Pangloss: Fast Entity Linking in Noisy Text Environments
Jul 16, 2018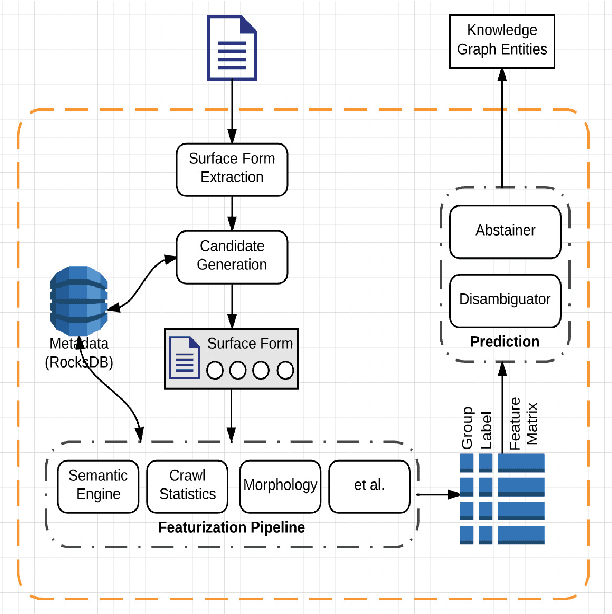
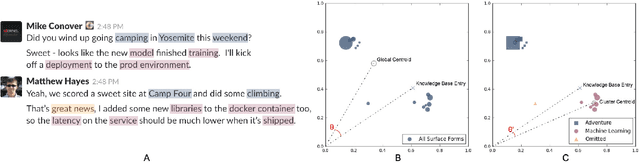
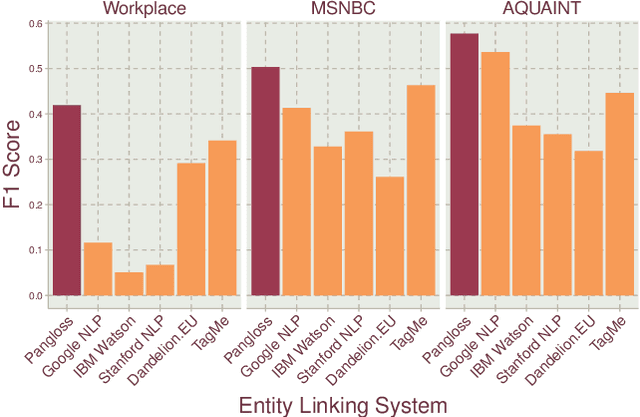
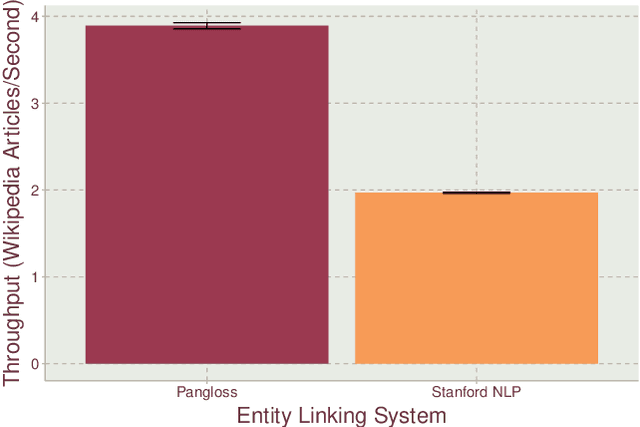
Abstract:Entity linking is the task of mapping potentially ambiguous terms in text to their constituent entities in a knowledge base like Wikipedia. This is useful for organizing content, extracting structured data from textual documents, and in machine learning relevance applications like semantic search, knowledge graph construction, and question answering. Traditionally, this work has focused on text that has been well-formed, like news articles, but in common real world datasets such as messaging, resumes, or short-form social media, non-grammatical, loosely-structured text adds a new dimension to this problem. This paper presents Pangloss, a production system for entity disambiguation on noisy text. Pangloss combines a probabilistic linear-time key phrase identification algorithm with a semantic similarity engine based on context-dependent document embeddings to achieve better than state-of-the-art results (>5% in F1) compared to other research or commercially available systems. In addition, Pangloss leverages a local embedded database with a tiered architecture to house its statistics and metadata, which allows rapid disambiguation in streaming contexts and on-device disambiguation in low-memory environments such as mobile phones.
A Linear Classifier Based on Entity Recognition Tools and a Statistical Approach to Method Extraction in the Protein-Protein Interaction Literature
Apr 22, 2011

Abstract:We participated, in the Article Classification and the Interaction Method subtasks (ACT and IMT, respectively) of the Protein-Protein Interaction task of the BioCreative III Challenge. For the ACT, we pursued an extensive testing of available Named Entity Recognition and dictionary tools, and used the most promising ones to extend our Variable Trigonometric Threshold linear classifier. For the IMT, we experimented with a primarily statistical approach, as opposed to employing a deeper natural language processing strategy. Finally, we also studied the benefits of integrating the method extraction approach that we have used for the IMT into the ACT pipeline. For the ACT, our linear article classifier leads to a ranking and classification performance significantly higher than all the reported submissions. For the IMT, our results are comparable to those of other systems, which took very different approaches. For the ACT, we show that the use of named entity recognition tools leads to a substantial improvement in the ranking and classification of articles relevant to protein-protein interaction. Thus, we show that our substantially expanded linear classifier is a very competitive classifier in this domain. Moreover, this classifier produces interpretable surfaces that can be understood as "rules" for human understanding of the classification. In terms of the IMT task, in contrast to other participants, our approach focused on identifying sentences that are likely to bear evidence for the application of a PPI detection method, rather than on classifying a document as relevant to a method. As BioCreative III did not perform an evaluation of the evidence provided by the system, we have conducted a separate assessment; the evaluators agree that our tool is indeed effective in detecting relevant evidence for PPI detection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge