Andrew Wong
Feature engineering workflow for activity recognition from synchronized inertial measurement units
Dec 18, 2019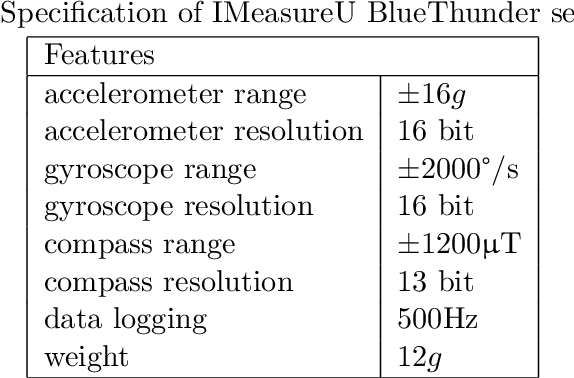
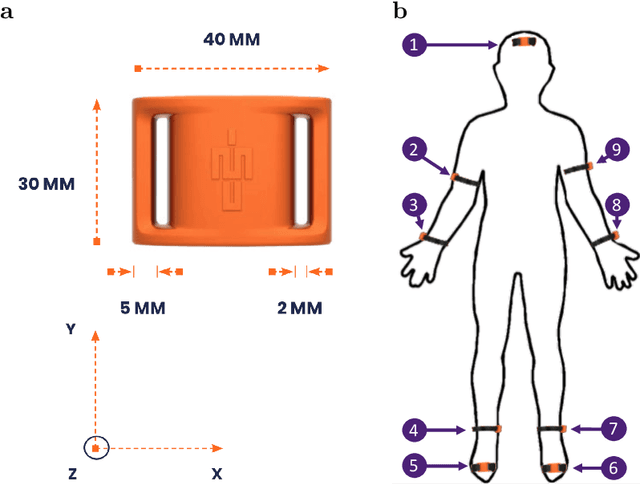
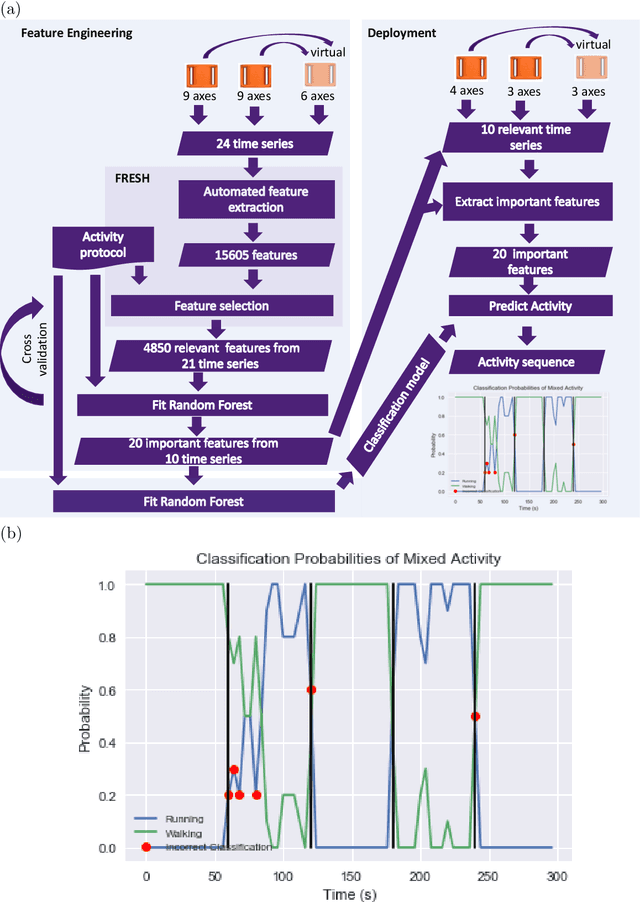
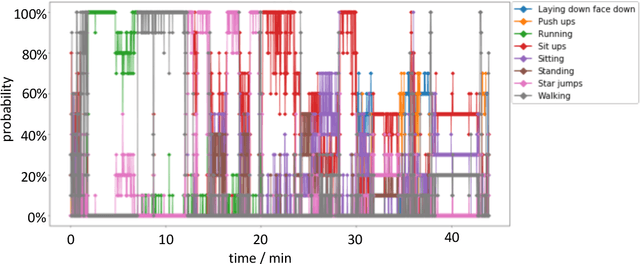
Abstract:The ubiquitous availability of wearable sensors is responsible for driving the Internet-of-Things but is also making an impact on sport sciences and precision medicine. While human activity recognition from smartphone data or other types of inertial measurement units (IMU) has evolved to one of the most prominent daily life examples of machine learning, the underlying process of time-series feature engineering still seems to be time-consuming. This lengthy process inhibits the development of IMU-based machine learning applications in sport science and precision medicine. This contribution discusses a feature engineering workflow, which automates the extraction of time-series feature on based on the FRESH algorithm (FeatuRe Extraction based on Scalable Hypothesis tests) to identify statistically significant features from synchronized IMU sensors (IMeasureU Ltd, NZ). The feature engineering workflow has five main steps: time-series engineering, automated time-series feature extraction, optimized feature extraction, fitting of a specialized classifier, and deployment of optimized machine learning pipeline. The workflow is discussed for the case of a user-specific running-walking classification, and the generalization to a multi-user multi-activity classification is demonstrated.
A Linear Classifier Based on Entity Recognition Tools and a Statistical Approach to Method Extraction in the Protein-Protein Interaction Literature
Apr 22, 2011

Abstract:We participated, in the Article Classification and the Interaction Method subtasks (ACT and IMT, respectively) of the Protein-Protein Interaction task of the BioCreative III Challenge. For the ACT, we pursued an extensive testing of available Named Entity Recognition and dictionary tools, and used the most promising ones to extend our Variable Trigonometric Threshold linear classifier. For the IMT, we experimented with a primarily statistical approach, as opposed to employing a deeper natural language processing strategy. Finally, we also studied the benefits of integrating the method extraction approach that we have used for the IMT into the ACT pipeline. For the ACT, our linear article classifier leads to a ranking and classification performance significantly higher than all the reported submissions. For the IMT, our results are comparable to those of other systems, which took very different approaches. For the ACT, we show that the use of named entity recognition tools leads to a substantial improvement in the ranking and classification of articles relevant to protein-protein interaction. Thus, we show that our substantially expanded linear classifier is a very competitive classifier in this domain. Moreover, this classifier produces interpretable surfaces that can be understood as "rules" for human understanding of the classification. In terms of the IMT task, in contrast to other participants, our approach focused on identifying sentences that are likely to bear evidence for the application of a PPI detection method, rather than on classifying a document as relevant to a method. As BioCreative III did not perform an evaluation of the evidence provided by the system, we have conducted a separate assessment; the evaluators agree that our tool is indeed effective in detecting relevant evidence for PPI detection methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge