Bruce W. Herr II
myAURA: Personalized health library for epilepsy management via knowledge graph sparsification and visualization
May 08, 2024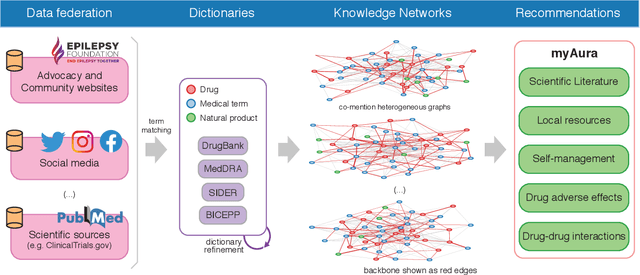
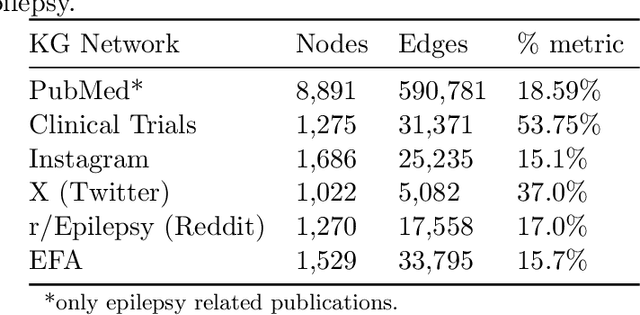
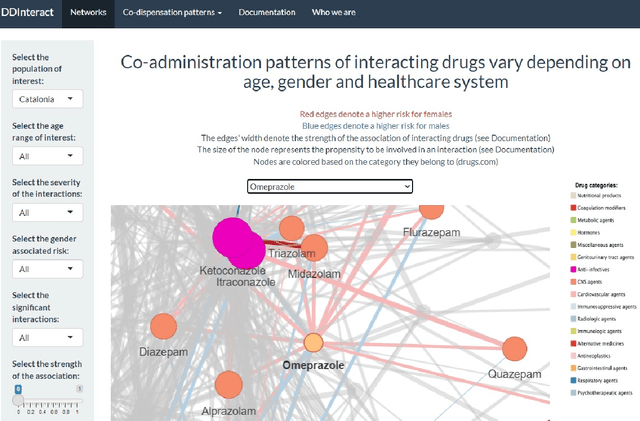
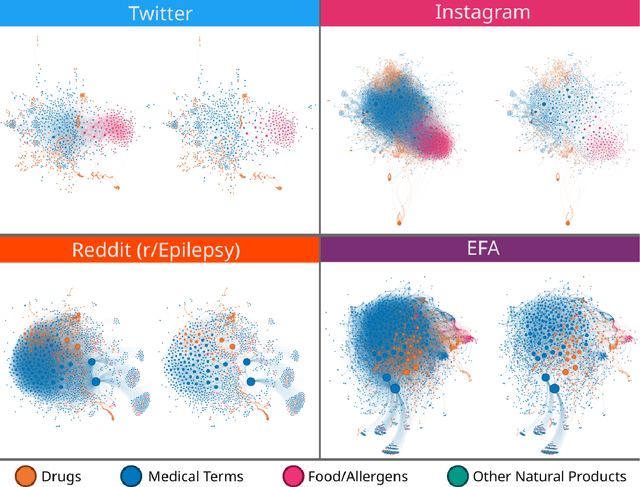
Abstract:Objective: We report the development of the patient-centered myAURA application and suite of methods designed to aid epilepsy patients, caregivers, and researchers in making decisions about care and self-management. Materials and Methods: myAURA rests on the federation of an unprecedented collection of heterogeneous data resources relevant to epilepsy, such as biomedical databases, social media, and electronic health records. A generalizable, open-source methodology was developed to compute a multi-layer knowledge graph linking all this heterogeneous data via the terms of a human-centered biomedical dictionary. Results: The power of the approach is first exemplified in the study of the drug-drug interaction phenomenon. Furthermore, we employ a novel network sparsification methodology using the metric backbone of weighted graphs, which reveals the most important edges for inference, recommendation, and visualization, such as pharmacology factors patients discuss on social media. The network sparsification approach also allows us to extract focused digital cohorts from social media whose discourse is more relevant to epilepsy or other biomedical problems. Finally, we present our patient-centered design and pilot-testing of myAURA, including its user interface, based on focus groups and other stakeholder input. Discussion: The ability to search and explore myAURA's heterogeneous data sources via a sparsified multi-layer knowledge graph, as well as the combination of those layers in a single map, are useful features for integrating relevant information for epilepsy. Conclusion: Our stakeholder-driven, scalable approach to integrate traditional and non-traditional data sources, enables biomedical discovery and data-powered patient self-management in epilepsy, and is generalizable to other chronic conditions.
Construction and Usage of a Human Body Common Coordinate Framework Comprising Clinical, Semantic, and Spatial Ontologies
Jul 28, 2020



Abstract:The National Institutes of Health's (NIH) Human Biomolecular Atlas Program (HuBMAP) aims to create a comprehensive high-resolution atlas of all the cells in the healthy human body. Multiple laboratories across the United States are collecting tissue specimens from different organs of donors who vary in sex, age, and body size. Integrating and harmonizing the data derived from these samples and 'mapping' them into a common three-dimensional (3D) space is a major challenge. The key to making this possible is a 'Common Coordinate Framework' (CCF), which provides a semantically annotated, 3D reference system for the entire body. The CCF enables contributors to HuBMAP to 'register' specimens and datasets within a common spatial reference system, and it supports a standardized way to query and 'explore' data in a spatially and semantically explicit manner. [...] This paper describes the construction and usage of a CCF for the human body and its reference implementation in HuBMAP. The CCF consists of (1) a CCF Clinical Ontology, which provides metadata about the specimen and donor (the 'who'); (2) a CCF Semantic Ontology, which describes 'what' part of the body a sample came from and details anatomical structures, cell types, and biomarkers (ASCT+B); and (3) a CCF Spatial Ontology, which indicates 'where' a tissue sample is located in a 3D coordinate system. An initial version of all three CCF ontologies has been implemented for the first HuBMAP Portal release. It was successfully used by Tissue Mapping Centers to semantically annotate and spatially register 48 kidney and spleen tissue blocks. The blocks can be queried and explored in their clinical, semantic, and spatial context via the CCF user interface in the HuBMAP Portal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge