Katy Börner
Department of Intelligent Systems Engineering, Indiana University, Bloomington, IN, USA
The IBEX Imaging Knowledge-Base: A Community Resource Enabling Adoption and Development of Immunofluoresence Imaging Methods
Dec 17, 2024Abstract:The iterative bleaching extends multiplexity (IBEX) Knowledge-Base is a central portal for researchers adopting IBEX and related 2D and 3D immunofluorescence imaging methods. The design of the Knowledge-Base is modeled after efforts in the open-source software community and includes three facets: a development platform (GitHub), static website, and service for data archiving. The Knowledge-Base facilitates the practice of open science throughout the research life cycle by providing validation data for recommended and non-recommended reagents, e.g., primary and secondary antibodies. In addition to reporting negative data, the Knowledge-Base empowers method adoption and evolution by providing a venue for sharing protocols, videos, datasets, software, and publications. A dedicated discussion forum fosters a sense of community among researchers while addressing questions not covered in published manuscripts. Together, scientists from around the world are advancing scientific discovery at a faster pace, reducing wasted time and effort, and instilling greater confidence in the resulting data.
myAURA: Personalized health library for epilepsy management via knowledge graph sparsification and visualization
May 08, 2024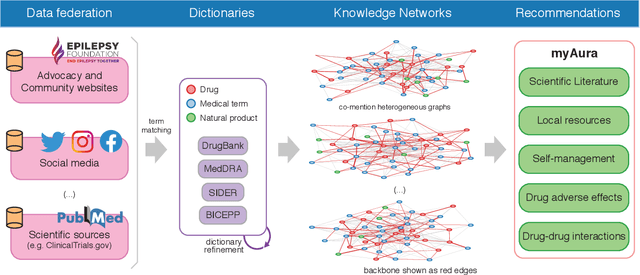
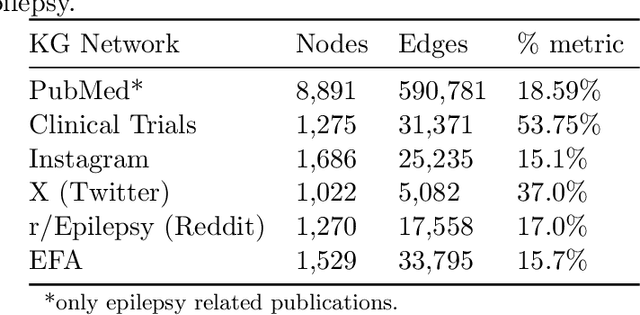
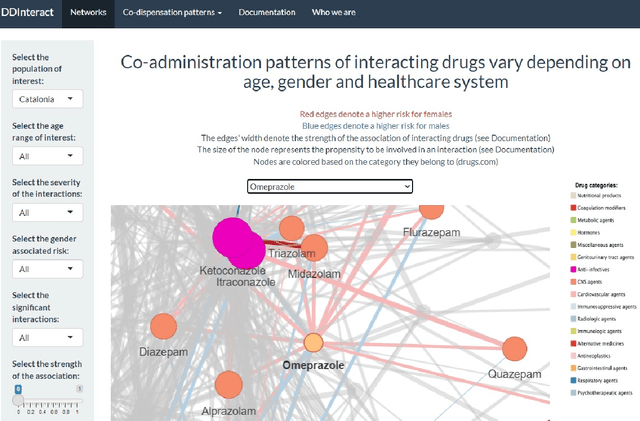
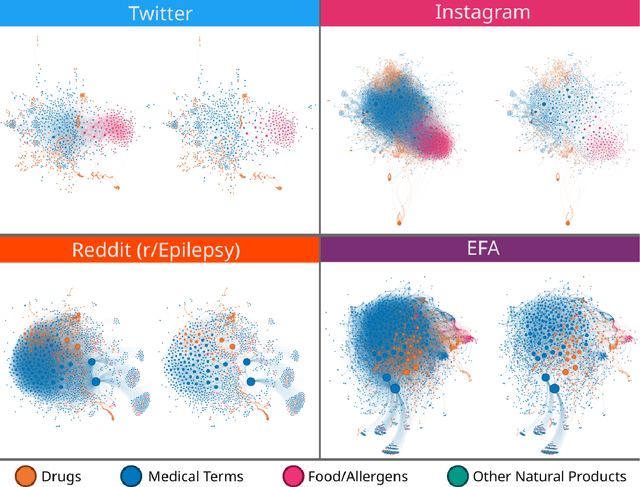
Abstract:Objective: We report the development of the patient-centered myAURA application and suite of methods designed to aid epilepsy patients, caregivers, and researchers in making decisions about care and self-management. Materials and Methods: myAURA rests on the federation of an unprecedented collection of heterogeneous data resources relevant to epilepsy, such as biomedical databases, social media, and electronic health records. A generalizable, open-source methodology was developed to compute a multi-layer knowledge graph linking all this heterogeneous data via the terms of a human-centered biomedical dictionary. Results: The power of the approach is first exemplified in the study of the drug-drug interaction phenomenon. Furthermore, we employ a novel network sparsification methodology using the metric backbone of weighted graphs, which reveals the most important edges for inference, recommendation, and visualization, such as pharmacology factors patients discuss on social media. The network sparsification approach also allows us to extract focused digital cohorts from social media whose discourse is more relevant to epilepsy or other biomedical problems. Finally, we present our patient-centered design and pilot-testing of myAURA, including its user interface, based on focus groups and other stakeholder input. Discussion: The ability to search and explore myAURA's heterogeneous data sources via a sparsified multi-layer knowledge graph, as well as the combination of those layers in a single map, are useful features for integrating relevant information for epilepsy. Conclusion: Our stakeholder-driven, scalable approach to integrate traditional and non-traditional data sources, enables biomedical discovery and data-powered patient self-management in epilepsy, and is generalizable to other chronic conditions.
Leveraging Online Shopping Behaviors as a Proxy for Personal Lifestyle Choices: New Insights into Chronic Disease Prevention Literacy
Apr 30, 2021



Abstract:Ubiquitous internet access is reshaping the way we live, but it is accompanied by unprecedented challenges to prevent chronic diseases planted in long exposure to unhealthy lifestyles. This paper proposes leveraging online shopping behaviors as a proxy for personal lifestyle choices to freshen chronic disease prevention literacy targeted for times when e-commerce user experience has been assimilated into most people's daily life. Here, retrospective longitudinal query logs and purchase records from millions of online shoppers were accessed, constructing a broad spectrum of lifestyle features covering assorted product categories and buyer personas. Using the lifestyle-related information preceding their first purchases of prescription drugs, we could determine associations between online shoppers' past lifestyle choices and if they suffered from a particular chronic disease. Novel lifestyle risk factors were discovered in two exemplars -- depression and diabetes, most of which showed cognitive congruence with existing healthcare knowledge. Further, such empirical findings could be adopted to locate online shoppers at high risk of chronic diseases with fair accuracy (e.g., [area under the receiver operating characteristic curve] AUC=0.68 for depression and AUC=0.70 for diabetes), closely matching the performance of screening surveys benchmarked against medical diagnosis. Unobtrusive chronic disease surveillance via e-commerce sites may soon meet consenting individuals in the digital space they already inhabit.
Construction and Usage of a Human Body Common Coordinate Framework Comprising Clinical, Semantic, and Spatial Ontologies
Jul 28, 2020



Abstract:The National Institutes of Health's (NIH) Human Biomolecular Atlas Program (HuBMAP) aims to create a comprehensive high-resolution atlas of all the cells in the healthy human body. Multiple laboratories across the United States are collecting tissue specimens from different organs of donors who vary in sex, age, and body size. Integrating and harmonizing the data derived from these samples and 'mapping' them into a common three-dimensional (3D) space is a major challenge. The key to making this possible is a 'Common Coordinate Framework' (CCF), which provides a semantically annotated, 3D reference system for the entire body. The CCF enables contributors to HuBMAP to 'register' specimens and datasets within a common spatial reference system, and it supports a standardized way to query and 'explore' data in a spatially and semantically explicit manner. [...] This paper describes the construction and usage of a CCF for the human body and its reference implementation in HuBMAP. The CCF consists of (1) a CCF Clinical Ontology, which provides metadata about the specimen and donor (the 'who'); (2) a CCF Semantic Ontology, which describes 'what' part of the body a sample came from and details anatomical structures, cell types, and biomarkers (ASCT+B); and (3) a CCF Spatial Ontology, which indicates 'where' a tissue sample is located in a 3D coordinate system. An initial version of all three CCF ontologies has been implemented for the first HuBMAP Portal release. It was successfully used by Tissue Mapping Centers to semantically annotate and spatially register 48 kidney and spleen tissue blocks. The blocks can be queried and explored in their clinical, semantic, and spatial context via the CCF user interface in the HuBMAP Portal.
Multi-level computational methods for interdisciplinary research in the HathiTrust Digital Library
Jun 08, 2017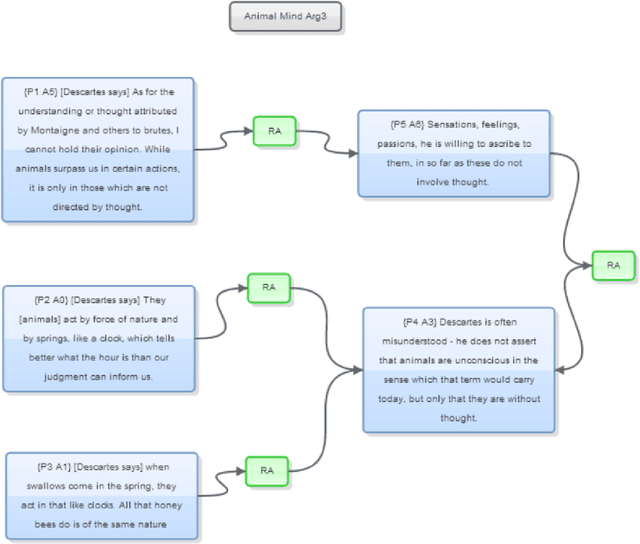
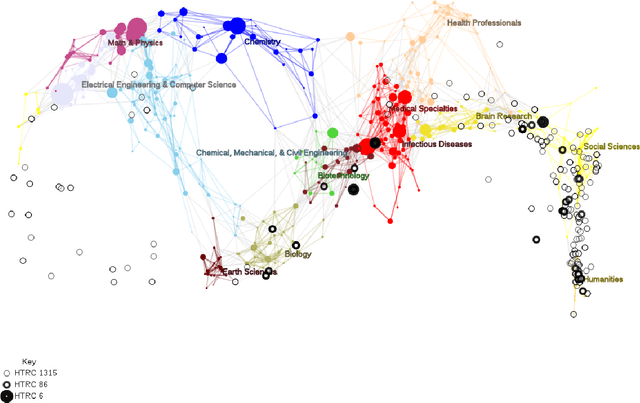

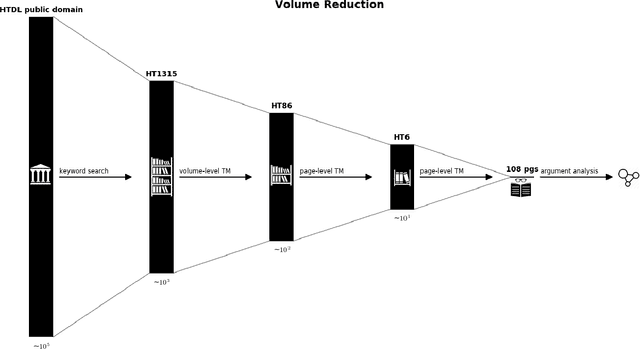
Abstract:We show how faceted search using a combination of traditional classification systems and mixed-membership topic models can go beyond keyword search to inform resource discovery, hypothesis formulation, and argument extraction for interdisciplinary research. Our test domain is the history and philosophy of scientific work on animal mind and cognition. The methods can be generalized to other research areas and ultimately support a system for semi-automatic identification of argument structures. We provide a case study for the application of the methods to the problem of identifying and extracting arguments about anthropomorphism during a critical period in the development of comparative psychology. We show how a combination of classification systems and mixed-membership models trained over large digital libraries can inform resource discovery in this domain. Through a novel approach of "drill-down" topic modeling---simultaneously reducing both the size of the corpus and the unit of analysis---we are able to reduce a large collection of fulltext volumes to a much smaller set of pages within six focal volumes containing arguments of interest to historians and philosophers of comparative psychology. The volumes identified in this way did not appear among the first ten results of the keyword search in the HathiTrust digital library and the pages bear the kind of "close reading" needed to generate original interpretations that is the heart of scholarly work in the humanities. Zooming back out, we provide a way to place the books onto a map of science originally constructed from very different data and for different purposes. The multilevel approach advances understanding of the intellectual and societal contexts in which writings are interpreted.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge