Ziv Yaniv
Bioinformatics and Computational Bioscience Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA
The IBEX Imaging Knowledge-Base: A Community Resource Enabling Adoption and Development of Immunofluoresence Imaging Methods
Dec 17, 2024Abstract:The iterative bleaching extends multiplexity (IBEX) Knowledge-Base is a central portal for researchers adopting IBEX and related 2D and 3D immunofluorescence imaging methods. The design of the Knowledge-Base is modeled after efforts in the open-source software community and includes three facets: a development platform (GitHub), static website, and service for data archiving. The Knowledge-Base facilitates the practice of open science throughout the research life cycle by providing validation data for recommended and non-recommended reagents, e.g., primary and secondary antibodies. In addition to reporting negative data, the Knowledge-Base empowers method adoption and evolution by providing a venue for sharing protocols, videos, datasets, software, and publications. A dedicated discussion forum fosters a sense of community among researchers while addressing questions not covered in published manuscripts. Together, scientists from around the world are advancing scientific discovery at a faster pace, reducing wasted time and effort, and instilling greater confidence in the resulting data.
A Distance Map Regularized CNN for Cardiac Cine MR Image Segmentation
Jan 04, 2019
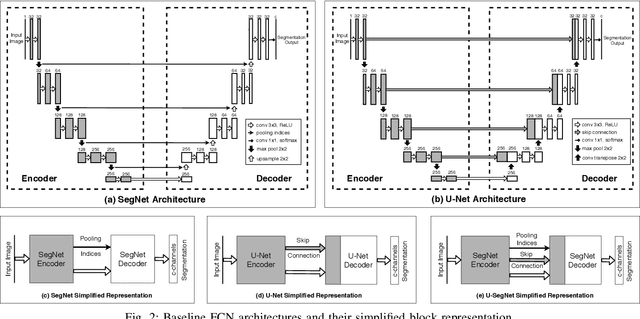
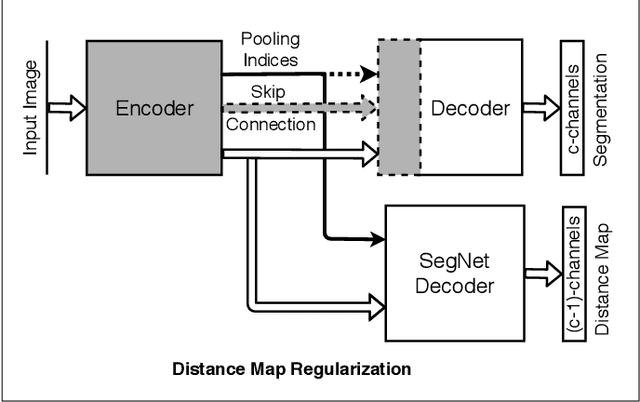
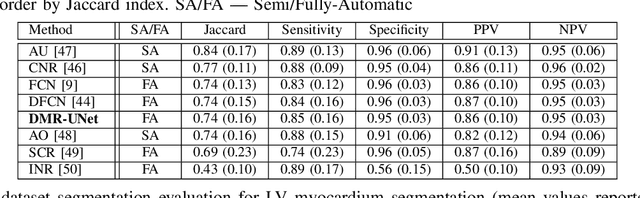
Abstract:Cardiac image segmentation is a critical process for generating personalized models of the heart and for quantifying cardiac performance parameters. Several convolutional neural network (CNN) architectures have been proposed to segment the heart chambers from cardiac cine MR images. Here we propose a multi-task learning (MTL)-based regularization framework for cardiac MR image segmentation. The network is trained to perform the main task of semantic segmentation, along with a simultaneous, auxiliary task of pixel-wise distance map regression. The proposed distance map regularizer is a decoder network added to the bottleneck layer of an existing CNN architecture, facilitating the network to learn robust global features. The regularizer block is removed after training, so that the original number of network parameters does not change. We show that the proposed regularization method improves both binary and multi-class segmentation performance over the corresponding state-of-the-art CNN architectures on two publicly available cardiac cine MRI datasets, obtaining average dice coefficient of 0.84$\pm$0.03 and 0.91$\pm$0.04, respectively. Furthermore, we also demonstrate improved generalization performance of the distance map regularized network on cross-dataset segmentation, showing as much as 41% improvement in average Dice coefficient from 0.57$\pm$0.28 to 0.80$\pm$0.13.
Left Ventricle Segmentation and Quantification from Cardiac Cine MR Images via Multi-task Learning
Sep 26, 2018



Abstract:Segmentation of the left ventricle and quantification of various cardiac contractile functions is crucial for the timely diagnosis and treatment of cardiovascular diseases. Traditionally, the two tasks have been tackled independently. Here we propose a convolutional neural network based multi-task learning approach to perform both tasks simultaneously, such that, the network learns better representation of the data with improved generalization performance. Probabilistic formulation of the problem enables learning the task uncertainties during the training, which are used to automatically compute the weights for the tasks. We performed a five fold cross-validation of the myocardium segmentation obtained from the proposed multi-task network on 97 patient 4-dimensional cardiac cine-MRI datasets available through the STACOM LV segmentation challenge against the provided gold-standard myocardium segmentation, obtaining a Dice overlap of $0.849 \pm 0.036$ and mean surface distance of $0.274 \pm 0.083$ mm, while simultaneously estimating the myocardial area with mean absolute difference error of $205\pm198$ mm$^2$.
Scalable Simple Linear Iterative Clustering (SSLIC) Using a Generic and Parallel Approach
Jul 23, 2018



Abstract:Superpixel algorithms have proven to be a useful initial step for segmentation and subsequent processing of images, reducing computational complexity by replacing the use of expensive per-pixel primitives with a higher-level abstraction, superpixels. They have been successfully applied both in the context of traditional image analysis and deep learning based approaches. In this work, we present a generalized implementation of the simple linear iterative clustering (SLIC) superpixel algorithm that has been generalized for n-dimensional scalar and multi-channel images. Additionally, the standard iterative implementation is replaced by a parallel, multi-threaded one. We describe the implementation details and analyze its scalability using a strong scaling formulation. Quantitative evaluation is performed using a 3D image, the Visible Human cryosection dataset, and a 2D image from the same dataset. Results show good scalability with runtime gains even when using a large number of threads that exceeds the physical number of available cores (hyperthreading).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge