Aehong Min
Refinement of an Epilepsy Dictionary through Human Annotation of Health-related posts on Instagram
May 14, 2024Abstract:We used a dictionary built from biomedical terminology extracted from various sources such as DrugBank, MedDRA, MedlinePlus, TCMGeneDIT, to tag more than 8 million Instagram posts by users who have mentioned an epilepsy-relevant drug at least once, between 2010 and early 2016. A random sample of 1,771 posts with 2,947 term matches was evaluated by human annotators to identify false-positives. OpenAI's GPT series models were compared against human annotation. Frequent terms with a high false-positive rate were removed from the dictionary. Analysis of the estimated false-positive rates of the annotated terms revealed 8 ambiguous terms (plus synonyms) used in Instagram posts, which were removed from the original dictionary. To study the effect of removing those terms, we constructed knowledge networks using the refined and the original dictionaries and performed an eigenvector-centrality analysis on both networks. We show that the refined dictionary thus produced leads to a significantly different rank of important terms, as measured by their eigenvector-centrality of the knowledge networks. Furthermore, the most important terms obtained after refinement are of greater medical relevance. In addition, we show that OpenAI's GPT series models fare worse than human annotators in this task.
myAURA: Personalized health library for epilepsy management via knowledge graph sparsification and visualization
May 08, 2024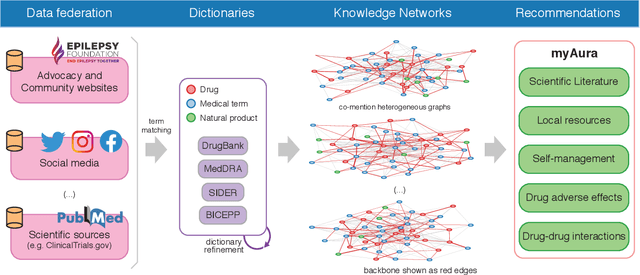
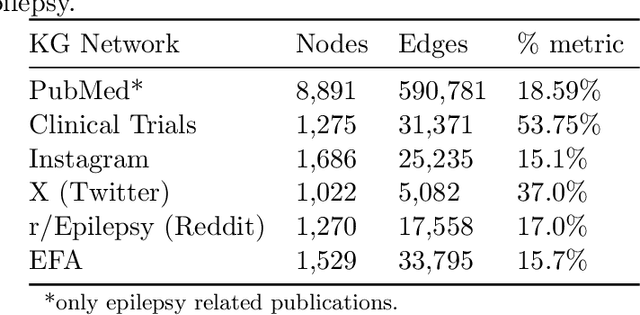
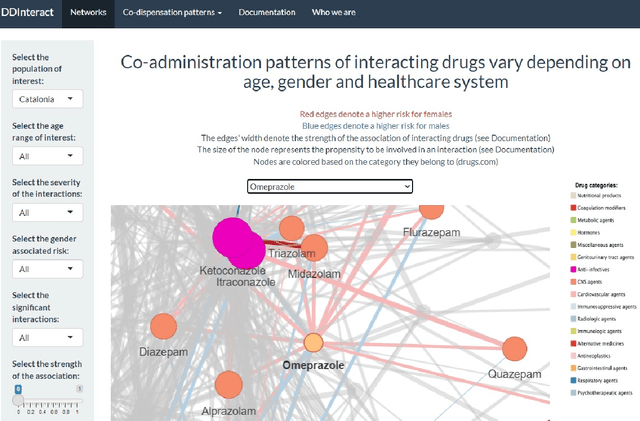
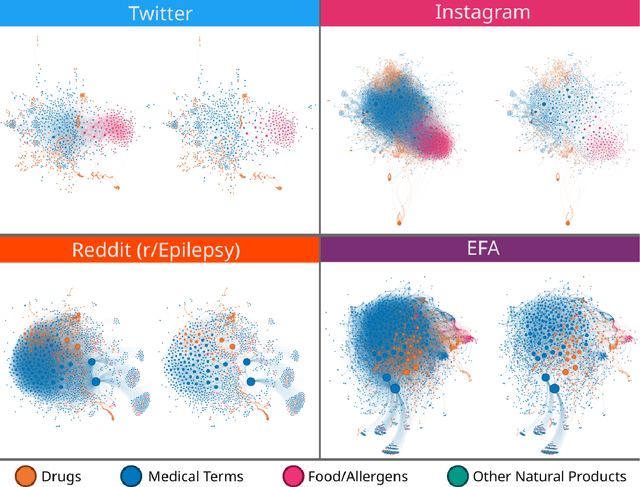
Abstract:Objective: We report the development of the patient-centered myAURA application and suite of methods designed to aid epilepsy patients, caregivers, and researchers in making decisions about care and self-management. Materials and Methods: myAURA rests on the federation of an unprecedented collection of heterogeneous data resources relevant to epilepsy, such as biomedical databases, social media, and electronic health records. A generalizable, open-source methodology was developed to compute a multi-layer knowledge graph linking all this heterogeneous data via the terms of a human-centered biomedical dictionary. Results: The power of the approach is first exemplified in the study of the drug-drug interaction phenomenon. Furthermore, we employ a novel network sparsification methodology using the metric backbone of weighted graphs, which reveals the most important edges for inference, recommendation, and visualization, such as pharmacology factors patients discuss on social media. The network sparsification approach also allows us to extract focused digital cohorts from social media whose discourse is more relevant to epilepsy or other biomedical problems. Finally, we present our patient-centered design and pilot-testing of myAURA, including its user interface, based on focus groups and other stakeholder input. Discussion: The ability to search and explore myAURA's heterogeneous data sources via a sparsified multi-layer knowledge graph, as well as the combination of those layers in a single map, are useful features for integrating relevant information for epilepsy. Conclusion: Our stakeholder-driven, scalable approach to integrate traditional and non-traditional data sources, enables biomedical discovery and data-powered patient self-management in epilepsy, and is generalizable to other chronic conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge