You Chen
LexEval: A Comprehensive Chinese Legal Benchmark for Evaluating Large Language Models
Sep 30, 2024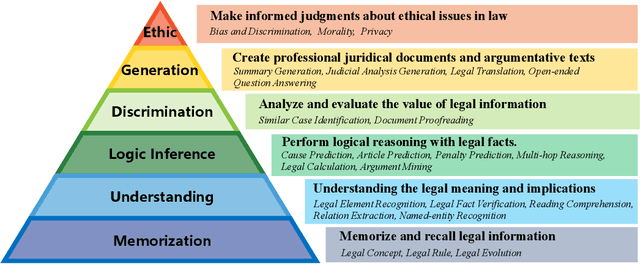



Abstract:Large language models (LLMs) have made significant progress in natural language processing tasks and demonstrate considerable potential in the legal domain. However, legal applications demand high standards of accuracy, reliability, and fairness. Applying existing LLMs to legal systems without careful evaluation of their potential and limitations could pose significant risks in legal practice. To this end, we introduce a standardized comprehensive Chinese legal benchmark LexEval. This benchmark is notable in the following three aspects: (1) Ability Modeling: We propose a new taxonomy of legal cognitive abilities to organize different tasks. (2) Scale: To our knowledge, LexEval is currently the largest Chinese legal evaluation dataset, comprising 23 tasks and 14,150 questions. (3) Data: we utilize formatted existing datasets, exam datasets and newly annotated datasets by legal experts to comprehensively evaluate the various capabilities of LLMs. LexEval not only focuses on the ability of LLMs to apply fundamental legal knowledge but also dedicates efforts to examining the ethical issues involved in their application. We evaluated 38 open-source and commercial LLMs and obtained some interesting findings. The experiments and findings offer valuable insights into the challenges and potential solutions for developing Chinese legal systems and LLM evaluation pipelines. The LexEval dataset and leaderboard are publicly available at \url{https://github.com/CSHaitao/LexEval} and will be continuously updated.
ECRTime: Ensemble Integration of Classification and Retrieval for Time Series Classification
Jul 20, 2024



Abstract:Deep learning-based methods for Time Series Classification (TSC) typically utilize deep networks to extract features, which are then processed through a combination of a Fully Connected (FC) layer and a SoftMax function. However, we have observed the phenomenon of inter-class similarity and intra-class inconsistency in the datasets from the UCR archive and further analyzed how this phenomenon adversely affects the "FC+SoftMax" paradigm. To address the issue, we introduce ECR, which, for the first time to our knowledge, applies deep learning-based retrieval algorithm to the TSC problem and integrates classification and retrieval models. Experimental results on 112 UCR datasets demonstrate that ECR is state-of-the-art(sota) compared to existing deep learning-based methods. Furthermore, we have developed a more precise classifier, ECRTime, which is an ensemble of ECR. ECRTime surpasses the currently most accurate deep learning classifier, InceptionTime, in terms of accuracy, achieving this with reduced training time and comparable scalability.
Towards an In-Depth Comprehension of Case Relevance for Better Legal Retrieval
Apr 01, 2024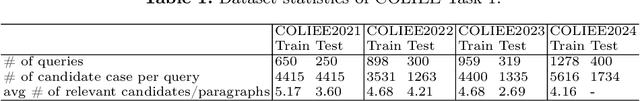


Abstract:Legal retrieval techniques play an important role in preserving the fairness and equality of the judicial system. As an annually well-known international competition, COLIEE aims to advance the development of state-of-the-art retrieval models for legal texts. This paper elaborates on the methodology employed by the TQM team in COLIEE2024.Specifically, we explored various lexical matching and semantic retrieval models, with a focus on enhancing the understanding of case relevance. Additionally, we endeavor to integrate various features using the learning-to-rank technique. Furthermore, fine heuristic pre-processing and post-processing methods have been proposed to mitigate irrelevant information. Consequently, our methodology achieved remarkable performance in COLIEE2024, securing first place in Task 1 and third place in Task 3. We anticipate that our proposed approach can contribute valuable insights to the advancement of legal retrieval technology.
Thinking about GPT-3 In-Context Learning for Biomedical IE? Think Again
Mar 16, 2022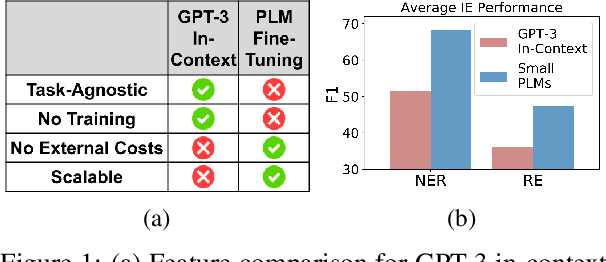
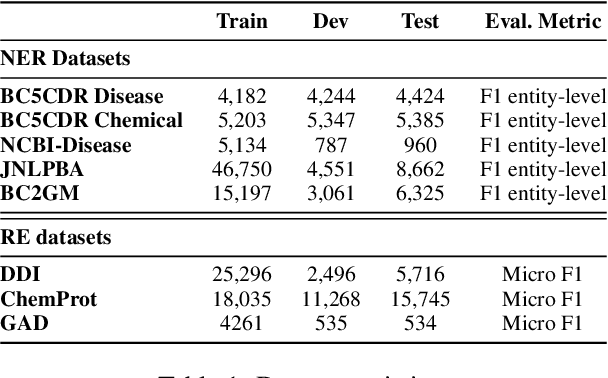

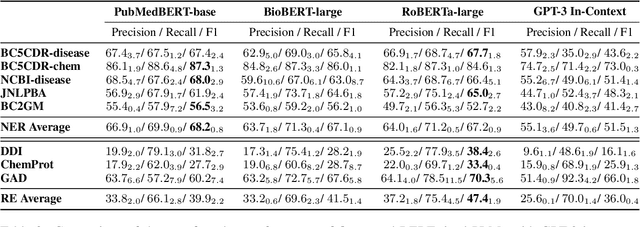
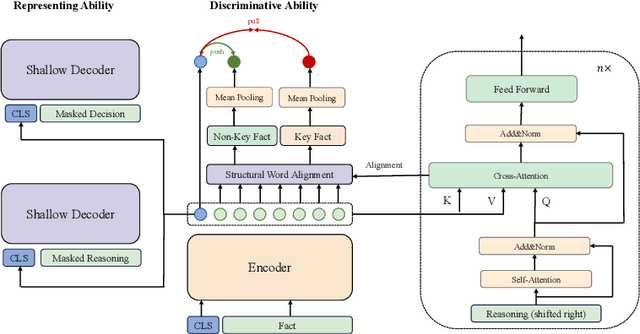
Abstract:The strong few-shot in-context learning capability of large pre-trained language models (PLMs) such as GPT-3 is highly appealing for biomedical applications where data annotation is particularly costly. In this paper, we present the first systematic and comprehensive study to compare the few-shot performance of GPT-3 in-context learning with fine-tuning smaller (i.e., BERT-sized) PLMs on two highly representative biomedical information extraction tasks, named entity recognition and relation extraction. We follow the true few-shot setting to avoid overestimating models' few-shot performance by model selection over a large validation set. We also optimize GPT-3's performance with known techniques such as contextual calibration and dynamic in-context example retrieval. However, our results show that GPT-3 still significantly underperforms compared with simply fine-tuning a smaller PLM using the same small training set. Moreover, what is equally important for practical applications is that adding more labeled data would reliably yield an improvement in model performance. While that is the case when fine-tuning small PLMs, GPT-3's performance barely improves when adding more data. In-depth analyses further reveal issues of the in-context learning setting that may be detrimental to information extraction tasks in general. Given the high cost of experimenting with GPT-3, we hope our study provides guidance for biomedical researchers and practitioners towards more promising directions such as fine-tuning GPT-3 or small PLMs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge