Clay Washington
Thinking about GPT-3 In-Context Learning for Biomedical IE? Think Again
Mar 16, 2022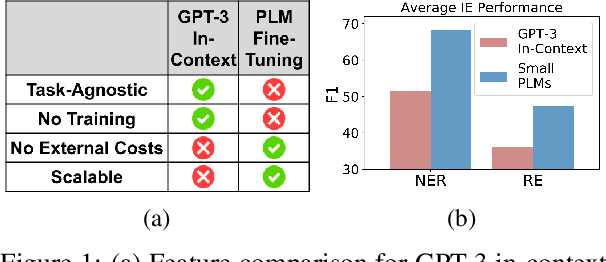
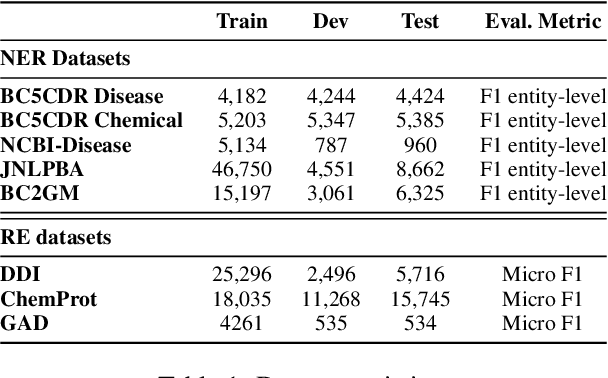

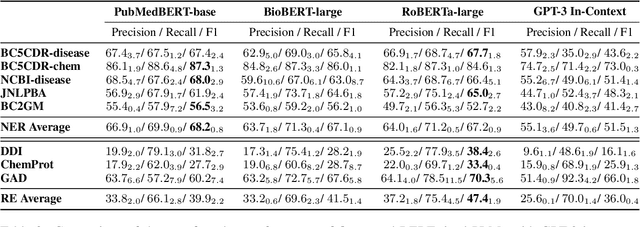
Abstract:The strong few-shot in-context learning capability of large pre-trained language models (PLMs) such as GPT-3 is highly appealing for biomedical applications where data annotation is particularly costly. In this paper, we present the first systematic and comprehensive study to compare the few-shot performance of GPT-3 in-context learning with fine-tuning smaller (i.e., BERT-sized) PLMs on two highly representative biomedical information extraction tasks, named entity recognition and relation extraction. We follow the true few-shot setting to avoid overestimating models' few-shot performance by model selection over a large validation set. We also optimize GPT-3's performance with known techniques such as contextual calibration and dynamic in-context example retrieval. However, our results show that GPT-3 still significantly underperforms compared with simply fine-tuning a smaller PLM using the same small training set. Moreover, what is equally important for practical applications is that adding more labeled data would reliably yield an improvement in model performance. While that is the case when fine-tuning small PLMs, GPT-3's performance barely improves when adding more data. In-depth analyses further reveal issues of the in-context learning setting that may be detrimental to information extraction tasks in general. Given the high cost of experimenting with GPT-3, we hope our study provides guidance for biomedical researchers and practitioners towards more promising directions such as fine-tuning GPT-3 or small PLMs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge