Bernal Jiménez Gutiérrez
Mind2Web 2: Evaluating Agentic Search with Agent-as-a-Judge
Jun 26, 2025Abstract:Agentic search such as Deep Research systems, where large language models autonomously browse the web, synthesize information, and return comprehensive citation-backed answers, represents a major shift in how users interact with web-scale information. While promising greater efficiency and cognitive offloading, the growing complexity and open-endedness of agentic search have outpaced existing evaluation benchmarks and methodologies, which largely assume short search horizons and static answers. In this paper, we introduce Mind2Web 2, a benchmark of 130 realistic, high-quality, and long-horizon tasks that require real-time web browsing and extensive information synthesis, constructed with over 1,000 hours of human labor. To address the challenge of evaluating time-varying and complex answers, we propose a novel Agent-as-a-Judge framework. Our method constructs task-specific judge agents based on a tree-structured rubric design to automatically assess both answer correctness and source attribution. We conduct a comprehensive evaluation of nine frontier agentic search systems and human performance, along with a detailed error analysis to draw insights for future development. The best-performing system, OpenAI Deep Research, can already achieve 50-70% of human performance while spending half the time, showing a great potential. Altogether, Mind2Web 2 provides a rigorous foundation for developing and benchmarking the next generation of agentic search systems.
From RAG to Memory: Non-Parametric Continual Learning for Large Language Models
Feb 20, 2025Abstract:Our ability to continuously acquire, organize, and leverage knowledge is a key feature of human intelligence that AI systems must approximate to unlock their full potential. Given the challenges in continual learning with large language models (LLMs), retrieval-augmented generation (RAG) has become the dominant way to introduce new information. However, its reliance on vector retrieval hinders its ability to mimic the dynamic and interconnected nature of human long-term memory. Recent RAG approaches augment vector embeddings with various structures like knowledge graphs to address some of these gaps, namely sense-making and associativity. However, their performance on more basic factual memory tasks drops considerably below standard RAG. We address this unintended deterioration and propose HippoRAG 2, a framework that outperforms standard RAG comprehensively on factual, sense-making, and associative memory tasks. HippoRAG 2 builds upon the Personalized PageRank algorithm used in HippoRAG and enhances it with deeper passage integration and more effective online use of an LLM. This combination pushes this RAG system closer to the effectiveness of human long-term memory, achieving a 7% improvement in associative memory tasks over the state-of-the-art embedding model while also exhibiting superior factual knowledge and sense-making memory capabilities. This work paves the way for non-parametric continual learning for LLMs. Our code and data will be released at https://github.com/OSU-NLP-Group/HippoRAG.
Attention in Large Language Models Yields Efficient Zero-Shot Re-Rankers
Oct 03, 2024Abstract:Information retrieval (IR) systems have played a vital role in modern digital life and have cemented their continued usefulness in this new era of generative AI via retrieval-augmented generation. With strong language processing capabilities and remarkable versatility, large language models (LLMs) have become popular choices for zero-shot re-ranking in IR systems. So far, LLM-based re-ranking methods rely on strong generative capabilities, which restricts their use to either specialized or powerful proprietary models. Given these restrictions, we ask: is autoregressive generation necessary and optimal for LLMs to perform re-ranking? We hypothesize that there are abundant signals relevant to re-ranking within LLMs that might not be used to their full potential via generation. To more directly leverage such signals, we propose in-context re-ranking (ICR), a novel method that leverages the change in attention pattern caused by the search query for accurate and efficient re-ranking. To mitigate the intrinsic biases in LLMs, we propose a calibration method using a content-free query. Due to the absence of generation, ICR only requires two ($O(1)$) forward passes to re-rank $N$ documents, making it substantially more efficient than generative re-ranking methods that require at least $O(N)$ forward passes. Our novel design also enables ICR to be applied to any LLM without specialized training while guaranteeing a well-formed ranking. Extensive experiments with two popular open-weight LLMs on standard single-hop and multi-hop information retrieval benchmarks show that ICR outperforms RankGPT while cutting the latency by more than 60% in practice. Through detailed analyses, we show that ICR's performance is specially strong on tasks that require more complex re-ranking signals. Our findings call for further exploration on novel ways of utilizing open-weight LLMs beyond text generation.
HippoRAG: Neurobiologically Inspired Long-Term Memory for Large Language Models
May 23, 2024Abstract:In order to thrive in hostile and ever-changing natural environments, mammalian brains evolved to store large amounts of knowledge about the world and continually integrate new information while avoiding catastrophic forgetting. Despite the impressive accomplishments, large language models (LLMs), even with retrieval-augmented generation (RAG), still struggle to efficiently and effectively integrate a large amount of new experiences after pre-training. In this work, we introduce HippoRAG, a novel retrieval framework inspired by the hippocampal indexing theory of human long-term memory to enable deeper and more efficient knowledge integration over new experiences. HippoRAG synergistically orchestrates LLMs, knowledge graphs, and the Personalized PageRank algorithm to mimic the different roles of neocortex and hippocampus in human memory. We compare HippoRAG with existing RAG methods on multi-hop question answering and show that our method outperforms the state-of-the-art methods remarkably, by up to 20%. Single-step retrieval with HippoRAG achieves comparable or better performance than iterative retrieval like IRCoT while being 10-30 times cheaper and 6-13 times faster, and integrating HippoRAG into IRCoT brings further substantial gains. Finally, we show that our method can tackle new types of scenarios that are out of reach of existing methods. Code and data are available at https://github.com/OSU-NLP-Group/HippoRAG.
Biomedical Language Models are Robust to Sub-optimal Tokenization
Jul 10, 2023



Abstract:As opposed to general English, many concepts in biomedical terminology have been designed in recent history by biomedical professionals with the goal of being precise and concise. This is often achieved by concatenating meaningful biomedical morphemes to create new semantic units. Nevertheless, most modern biomedical language models (LMs) are pre-trained using standard domain-specific tokenizers derived from large scale biomedical corpus statistics without explicitly leveraging the agglutinating nature of biomedical language. In this work, we first find that standard open-domain and biomedical tokenizers are largely unable to segment biomedical terms into meaningful components. Therefore, we hypothesize that using a tokenizer which segments biomedical terminology more accurately would enable biomedical LMs to improve their performance on downstream biomedical NLP tasks, especially ones which involve biomedical terms directly such as named entity recognition (NER) and entity linking. Surprisingly, we find that pre-training a biomedical LM using a more accurate biomedical tokenizer does not improve the entity representation quality of a language model as measured by several intrinsic and extrinsic measures such as masked language modeling prediction (MLM) accuracy as well as NER and entity linking performance. These quantitative findings, along with a case study which explores entity representation quality more directly, suggest that the biomedical pre-training process is quite robust to instances of sub-optimal tokenization.
Aligning Instruction Tasks Unlocks Large Language Models as Zero-Shot Relation Extractors
May 18, 2023Abstract:Recent work has shown that fine-tuning large language models (LLMs) on large-scale instruction-following datasets substantially improves their performance on a wide range of NLP tasks, especially in the zero-shot setting. However, even advanced instruction-tuned LLMs still fail to outperform small LMs on relation extraction (RE), a fundamental information extraction task. We hypothesize that instruction-tuning has been unable to elicit strong RE capabilities in LLMs due to RE's low incidence in instruction-tuning datasets, making up less than 1% of all tasks (Wang et al., 2022). To address this limitation, we propose QA4RE, a framework that aligns RE with question answering (QA), a predominant task in instruction-tuning datasets. Comprehensive zero-shot RE experiments over four datasets with two series of instruction-tuned LLMs (six LLMs in total) demonstrate that our QA4RE framework consistently improves LLM performance, strongly verifying our hypothesis and enabling LLMs to outperform strong zero-shot baselines by a large margin. Additionally, we provide thorough experiments and discussions to show the robustness, few-shot effectiveness, and strong transferability of our QA4RE framework. This work illustrates a promising way of adapting LLMs to challenging and underrepresented tasks by aligning these tasks with more common instruction-tuning tasks like QA.
Thinking about GPT-3 In-Context Learning for Biomedical IE? Think Again
Mar 16, 2022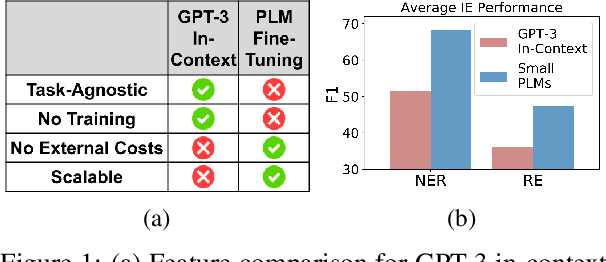
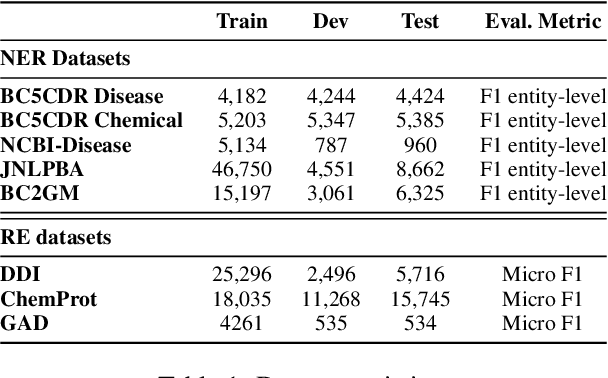

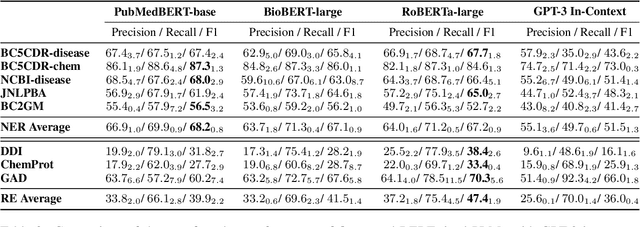
Abstract:The strong few-shot in-context learning capability of large pre-trained language models (PLMs) such as GPT-3 is highly appealing for biomedical applications where data annotation is particularly costly. In this paper, we present the first systematic and comprehensive study to compare the few-shot performance of GPT-3 in-context learning with fine-tuning smaller (i.e., BERT-sized) PLMs on two highly representative biomedical information extraction tasks, named entity recognition and relation extraction. We follow the true few-shot setting to avoid overestimating models' few-shot performance by model selection over a large validation set. We also optimize GPT-3's performance with known techniques such as contextual calibration and dynamic in-context example retrieval. However, our results show that GPT-3 still significantly underperforms compared with simply fine-tuning a smaller PLM using the same small training set. Moreover, what is equally important for practical applications is that adding more labeled data would reliably yield an improvement in model performance. While that is the case when fine-tuning small PLMs, GPT-3's performance barely improves when adding more data. In-depth analyses further reveal issues of the in-context learning setting that may be detrimental to information extraction tasks in general. Given the high cost of experimenting with GPT-3, we hope our study provides guidance for biomedical researchers and practitioners towards more promising directions such as fine-tuning GPT-3 or small PLMs.
Document Classification for COVID-19 Literature
Jun 15, 2020



Abstract:The global pandemic has made it more important than ever to quickly and accurately retrieve relevant scientific literature for effective consumption by researchers in a wide range of fields. We provide an analysis of several multi-label document classification models on the LitCovid dataset, a growing collection of 8,000 research papers regarding the novel 2019 coronavirus. We find that pre-trained language models fine-tuned on this dataset outperform all other baselines and that the BioBERT and novel Longformer models surpass all others with almost equivalent micro-F1 and accuracy scores of around 81% and 69% on the test set. We evaluate the data efficiency and generalizability of these models as essential features of any system prepared to deal with an urgent situation like the current health crisis. Finally, we explore 50 errors made by the best performing models on LitCovid documents and find that they often (1) correlate certain labels too closely together and (2) fail to focus on discriminative sections of the articles; both of which are important issues to address in future work. Both data and code are available on GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge