Jingkun Chen
VDNeRF: Vision-only Dynamic Neural Radiance Field for Urban Scenes
Nov 09, 2025Abstract:Neural Radiance Fields (NeRFs) implicitly model continuous three-dimensional scenes using a set of images with known camera poses, enabling the rendering of photorealistic novel views. However, existing NeRF-based methods encounter challenges in applications such as autonomous driving and robotic perception, primarily due to the difficulty of capturing accurate camera poses and limitations in handling large-scale dynamic environments. To address these issues, we propose Vision-only Dynamic NeRF (VDNeRF), a method that accurately recovers camera trajectories and learns spatiotemporal representations for dynamic urban scenes without requiring additional camera pose information or expensive sensor data. VDNeRF employs two separate NeRF models to jointly reconstruct the scene. The static NeRF model optimizes camera poses and static background, while the dynamic NeRF model incorporates the 3D scene flow to ensure accurate and consistent reconstruction of dynamic objects. To address the ambiguity between camera motion and independent object motion, we design an effective and powerful training framework to achieve robust camera pose estimation and self-supervised decomposition of static and dynamic elements in a scene. Extensive evaluations on mainstream urban driving datasets demonstrate that VDNeRF surpasses state-of-the-art NeRF-based pose-free methods in both camera pose estimation and dynamic novel view synthesis.
The best performance in the CARE 2025 -- Liver Task (LiSeg-Contrast): Contrast-Aware Semi-Supervised Segmentation with Domain Generalization and Test-Time Adaptation
Oct 05, 2025



Abstract:Accurate liver segmentation from contrast-enhanced MRI is essential for diagnosis, treatment planning, and disease monitoring. However, it remains challenging due to limited annotated data, heterogeneous enhancement protocols, and significant domain shifts across scanners and institutions. Traditional image-to-image translation frameworks have made great progress in domain generalization, but their application is not straightforward. For example, Pix2Pix requires image registration, and cycle-GAN cannot be integrated seamlessly into segmentation pipelines. Meanwhile, these methods are originally used to deal with cross-modality scenarios, and often introduce structural distortions and suffer from unstable training, which may pose drawbacks in our single-modality scenario. To address these challenges, we propose CoSSeg-TTA, a compact segmentation framework for the GED4 (Gd-EOB-DTPA enhanced hepatobiliary phase MRI) modality built upon nnU-Netv2 and enhanced with a semi-supervised mean teacher scheme to exploit large amounts of unlabeled volumes. A domain adaptation module, incorporating a randomized histogram-based style appearance transfer function and a trainable contrast-aware network, enriches domain diversity and mitigates cross-center variability. Furthermore, a continual test-time adaptation strategy is employed to improve robustness during inference. Extensive experiments demonstrate that our framework consistently outperforms the nnU-Netv2 baseline, achieving superior Dice score and Hausdorff Distance while exhibiting strong generalization to unseen domains under low-annotation conditions.
TABNet: A Triplet Augmentation Self-Recovery Framework with Boundary-Aware Pseudo-Labels for Medical Image Segmentation
Jul 03, 2025Abstract:Background and objective: Medical image segmentation is a core task in various clinical applications. However, acquiring large-scale, fully annotated medical image datasets is both time-consuming and costly. Scribble annotations, as a form of sparse labeling, provide an efficient and cost-effective alternative for medical image segmentation. However, the sparsity of scribble annotations limits the feature learning of the target region and lacks sufficient boundary supervision, which poses significant challenges for training segmentation networks. Methods: We propose TAB Net, a novel weakly-supervised medical image segmentation framework, consisting of two key components: the triplet augmentation self-recovery (TAS) module and the boundary-aware pseudo-label supervision (BAP) module. The TAS module enhances feature learning through three complementary augmentation strategies: intensity transformation improves the model's sensitivity to texture and contrast variations, cutout forces the network to capture local anatomical structures by masking key regions, and jigsaw augmentation strengthens the modeling of global anatomical layout by disrupting spatial continuity. By guiding the network to recover complete masks from diverse augmented inputs, TAS promotes a deeper semantic understanding of medical images under sparse supervision. The BAP module enhances pseudo-supervision accuracy and boundary modeling by fusing dual-branch predictions into a loss-weighted pseudo-label and introducing a boundary-aware loss for fine-grained contour refinement. Results: Experimental evaluations on two public datasets, ACDC and MSCMR seg, demonstrate that TAB Net significantly outperforms state-of-the-art methods for scribble-based weakly supervised segmentation. Moreover, it achieves performance comparable to that of fully supervised methods.
From Gaze to Insight: Bridging Human Visual Attention and Vision Language Model Explanation for Weakly-Supervised Medical Image Segmentation
Apr 15, 2025Abstract:Medical image segmentation remains challenging due to the high cost of pixel-level annotations for training. In the context of weak supervision, clinician gaze data captures regions of diagnostic interest; however, its sparsity limits its use for segmentation. In contrast, vision-language models (VLMs) provide semantic context through textual descriptions but lack the explanation precision required. Recognizing that neither source alone suffices, we propose a teacher-student framework that integrates both gaze and language supervision, leveraging their complementary strengths. Our key insight is that gaze data indicates where clinicians focus during diagnosis, while VLMs explain why those regions are significant. To implement this, the teacher model first learns from gaze points enhanced by VLM-generated descriptions of lesion morphology, establishing a foundation for guiding the student model. The teacher then directs the student through three strategies: (1) Multi-scale feature alignment to fuse visual cues with textual semantics; (2) Confidence-weighted consistency constraints to focus on reliable predictions; (3) Adaptive masking to limit error propagation in uncertain areas. Experiments on the Kvasir-SEG, NCI-ISBI, and ISIC datasets show that our method achieves Dice scores of 80.78%, 80.53%, and 84.22%, respectively-improving 3-5% over gaze baselines without increasing the annotation burden. By preserving correlations among predictions, gaze data, and lesion descriptions, our framework also maintains clinical interpretability. This work illustrates how integrating human visual attention with AI-generated semantic context can effectively overcome the limitations of individual weak supervision signals, thereby advancing the development of deployable, annotation-efficient medical AI systems. Code is available at: https://github.com/jingkunchen/FGI.git.
Unsupervised Patch-GAN with Targeted Patch Ranking for Fine-Grained Novelty Detection in Medical Imaging
Jan 29, 2025



Abstract:Detecting novel anomalies in medical imaging is challenging due to the limited availability of labeled data for rare abnormalities, which often display high variability and subtlety. This challenge is further compounded when small abnormal regions are embedded within larger normal areas, as whole-image predictions frequently overlook these subtle deviations. To address these issues, we propose an unsupervised Patch-GAN framework designed to detect and localize anomalies by capturing both local detail and global structure. Our framework first reconstructs masked images to learn fine-grained, normal-specific features, allowing for enhanced sensitivity to minor deviations from normality. By dividing these reconstructed images into patches and assessing the authenticity of each patch, our approach identifies anomalies at a more granular level, overcoming the limitations of whole-image evaluation. Additionally, a patch-ranking mechanism prioritizes regions with higher abnormal scores, reinforcing the alignment between local patch discrepancies and the global image context. Experimental results on the ISIC 2016 skin lesion and BraTS 2019 brain tumor datasets validate our framework's effectiveness, achieving AUCs of 95.79% and 96.05%, respectively, and outperforming three state-of-the-art baselines.
Contrast-Free Myocardial Scar Segmentation in Cine MRI using Motion and Texture Fusion
Jan 09, 2025Abstract:Late gadolinium enhancement MRI (LGE MRI) is the gold standard for the detection of myocardial scars for post myocardial infarction (MI). LGE MRI requires the injection of a contrast agent, which carries potential side effects and increases scanning time and patient discomfort. To address these issues, we propose a novel framework that combines cardiac motion observed in cine MRI with image texture information to segment the myocardium and scar tissue in the left ventricle. Cardiac motion tracking can be formulated as a full cardiac image cycle registration problem, which can be solved via deep neural networks. Experimental results prove that the proposed method can achieve scar segmentation based on non-contrasted cine images with comparable accuracy to LGE MRI. This demonstrates its potential as an alternative to contrast-enhanced techniques for scar detection.
HELPNet: Hierarchical Perturbations Consistency and Entropy-guided Ensemble for Scribble Supervised Medical Image Segmentation
Dec 25, 2024



Abstract:Creating fully annotated labels for medical image segmentation is prohibitively time-intensive and costly, emphasizing the necessity for innovative approaches that minimize reliance on detailed annotations. Scribble annotations offer a more cost-effective alternative, significantly reducing the expenses associated with full annotations. However, scribble annotations offer limited and imprecise information, failing to capture the detailed structural and boundary characteristics necessary for accurate organ delineation. To address these challenges, we propose HELPNet, a novel scribble-based weakly supervised segmentation framework, designed to bridge the gap between annotation efficiency and segmentation performance. HELPNet integrates three modules. The Hierarchical perturbations consistency (HPC) module enhances feature learning by employing density-controlled jigsaw perturbations across global, local, and focal views, enabling robust modeling of multi-scale structural representations. Building on this, the Entropy-guided pseudo-label (EGPL) module evaluates the confidence of segmentation predictions using entropy, generating high-quality pseudo-labels. Finally, the structural prior refinement (SPR) module incorporates connectivity and bounded priors to enhance the precision and reliability and pseudo-labels. Experimental results on three public datasets ACDC, MSCMRseg, and CHAOS show that HELPNet significantly outperforms state-of-the-art methods for scribble-based weakly supervised segmentation and achieves performance comparable to fully supervised methods. The code is available at https://github.com/IPMI-NWU/HELPNet.
Luminance Component Analysis for Exposure Correction
Nov 25, 2024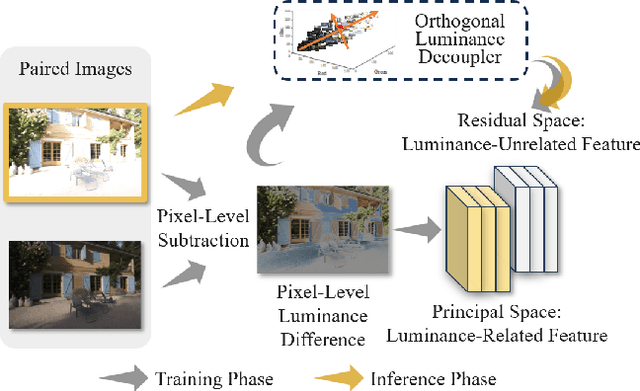

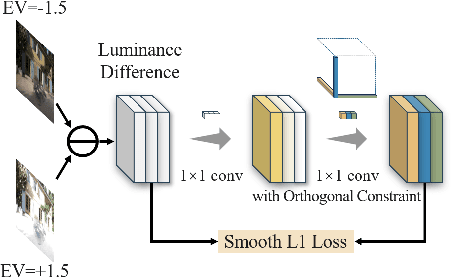

Abstract:Exposure correction methods aim to adjust the luminance while maintaining other luminance-unrelated information. However, current exposure correction methods have difficulty in fully separating luminance-related and luminance-unrelated components, leading to distortions in color, loss of detail, and requiring extra restoration procedures. Inspired by principal component analysis (PCA), this paper proposes an exposure correction method called luminance component analysis (LCA). LCA applies the orthogonal constraint to a U-Net structure to decouple luminance-related and luminance-unrelated features. With decoupled luminance-related features, LCA adjusts only the luminance-related components while keeping the luminance-unrelated components unchanged. To optimize the orthogonal constraint problem, LCA employs a geometric optimization algorithm, which converts the constrained problem in Euclidean space to an unconstrained problem in orthogonal Stiefel manifolds. Extensive experiments show that LCA can decouple the luminance feature from the RGB color space. Moreover, LCA achieves the best PSNR (21.33) and SSIM (0.88) in the exposure correction dataset with 28.72 FPS.
Deep Class-Specific Affinity-Guided Convolutional Network for Multimodal Unpaired Image Segmentation
Jan 05, 2021



Abstract:Multi-modal medical image segmentation plays an essential role in clinical diagnosis. It remains challenging as the input modalities are often not well-aligned spatially. Existing learning-based methods mainly consider sharing trainable layers across modalities and minimizing visual feature discrepancies. While the problem is often formulated as joint supervised feature learning, multiple-scale features and class-specific representation have not yet been explored. In this paper, we propose an affinity-guided fully convolutional network for multimodal image segmentation. To learn effective representations, we design class-specific affinity matrices to encode the knowledge of hierarchical feature reasoning, together with the shared convolutional layers to ensure the cross-modality generalization. Our affinity matrix does not depend on spatial alignments of the visual features and thus allows us to train with unpaired, multimodal inputs. We extensively evaluated our method on two public multimodal benchmark datasets and outperform state-of-the-art methods.
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020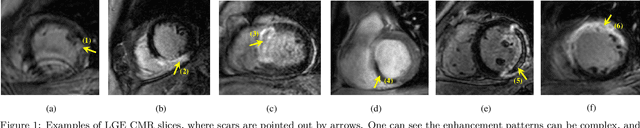
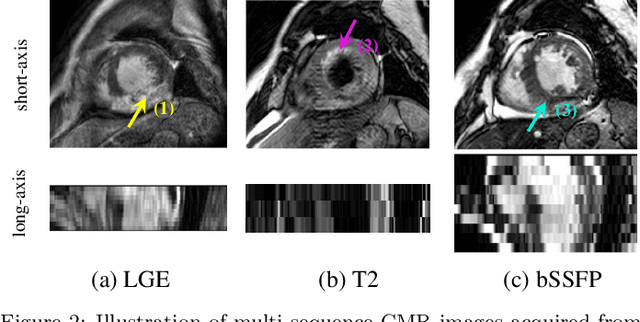
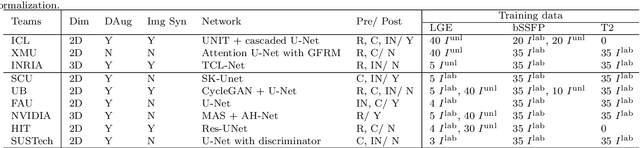
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge