Gaofeng Meng
UniSurg: A Video-Native Foundation Model for Universal Understanding of Surgical Videos
Feb 05, 2026Abstract:While foundation models have advanced surgical video analysis, current approaches rely predominantly on pixel-level reconstruction objectives that waste model capacity on low-level visual details - such as smoke, specular reflections, and fluid motion - rather than semantic structures essential for surgical understanding. We present UniSurg, a video-native foundation model that shifts the learning paradigm from pixel-level reconstruction to latent motion prediction. Built on the Video Joint Embedding Predictive Architecture (V-JEPA), UniSurg introduces three key technical innovations tailored to surgical videos: 1) motion-guided latent prediction to prioritize semantically meaningful regions, 2) spatiotemporal affinity self-distillation to enforce relational consistency, and 3) feature diversity regularization to prevent representation collapse in texture-sparse surgical scenes. To enable large-scale pretraining, we curate UniSurg-15M, the largest surgical video dataset to date, comprising 3,658 hours of video from 50 sources across 13 anatomical regions. Extensive experiments across 17 benchmarks demonstrate that UniSurg significantly outperforms state-of-the-art methods on surgical workflow recognition (+14.6% F1 on EgoSurgery, +10.3% on PitVis), action triplet recognition (39.54% mAP-IVT on CholecT50), skill assessment, polyp segmentation, and depth estimation. These results establish UniSurg as a new standard for universal, motion-oriented surgical video understanding.
VTCBench: Can Vision-Language Models Understand Long Context with Vision-Text Compression?
Dec 23, 2025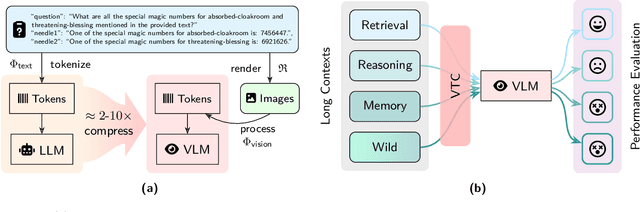

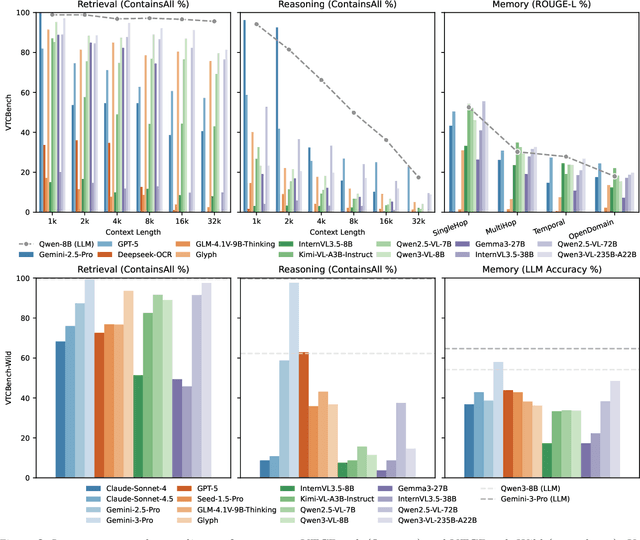
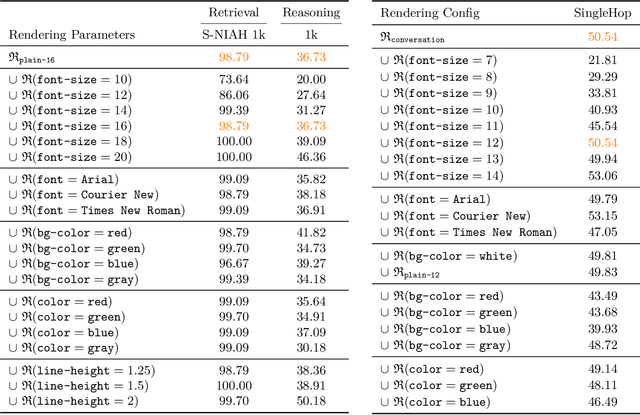
Abstract:The computational and memory overheads associated with expanding the context window of LLMs severely limit their scalability. A noteworthy solution is vision-text compression (VTC), exemplified by frameworks like DeepSeek-OCR and Glyph, which convert long texts into dense 2D visual representations, thereby achieving token compression ratios of 3x-20x. However, the impact of this high information density on the core long-context capabilities of vision-language models (VLMs) remains under-investigated. To address this gap, we introduce the first benchmark for VTC and systematically assess the performance of VLMs across three long-context understanding settings: VTC-Retrieval, which evaluates the model's ability to retrieve and aggregate information; VTC-Reasoning, which requires models to infer latent associations to locate facts with minimal lexical overlap; and VTC-Memory, which measures comprehensive question answering within long-term dialogue memory. Furthermore, we establish the VTCBench-Wild to simulate diverse input scenarios.We comprehensively evaluate leading open-source and proprietary models on our benchmarks. The results indicate that, despite being able to decode textual information (e.g., OCR) well, most VLMs exhibit a surprisingly poor long-context understanding ability with VTC-processed information, failing to capture long associations or dependencies in the context.This study provides a deep understanding of VTC and serves as a foundation for designing more efficient and scalable VLMs.
PDE-Agent: A toolchain-augmented multi-agent framework for PDE solving
Dec 22, 2025



Abstract:Solving Partial Differential Equations (PDEs) is a cornerstone of engineering and scientific research. Traditional methods for PDE solving are cumbersome, relying on manual setup and domain expertise. While Physics-Informed Neural Network (PINNs) introduced end-to-end neural network-based solutions, and frameworks like DeepXDE further enhanced automation, these approaches still depend on expert knowledge and lack full autonomy. In this work, we frame PDE solving as tool invocation via LLM-driven agents and introduce PDE-Agent, the first toolchain-augmented multi-agent collaboration framework, inheriting the reasoning capacity of LLMs and the controllability of external tools and enabling automated PDE solving from natural language descriptions. PDE-Agent leverages the strengths of multi-agent and multi-tool collaboration through two key innovations: (1) A Prog-Act framework with graph memory for multi-agent collaboration, which enables effective dynamic planning and error correction via dual-loop mechanisms (localized fixes and global revisions). (2) A Resource-Pool integrated with a tool-parameter separation mechanism for multi-tool collaboration. This centralizes the management of runtime artifacts and resolves inter-tool dependency gaps in existing frameworks. To validate and evaluate this new paradigm for PDE solving , we develop PDE-Bench, a multi-type PDE Benchmark for agent-based tool collaborative solving, and propose multi-level metrics for assessing tool coordination. Evaluations verify that PDE-Agent exhibits superior applicability and performance in complex multi-step, cross-step dependent tasks. This new paradigm of toolchain-augmented multi-agent PDE solving will further advance future developments in automated scientific computing. Our source code and dataset will be made publicly available.
PADReg: Physics-Aware Deformable Registration Guided by Contact Force for Ultrasound Sequences
Aug 12, 2025Abstract:Ultrasound deformable registration estimates spatial transformations between pairs of deformed ultrasound images, which is crucial for capturing biomechanical properties and enhancing diagnostic accuracy in diseases such as thyroid nodules and breast cancer. However, ultrasound deformable registration remains highly challenging, especially under large deformation. The inherently low contrast, heavy noise and ambiguous tissue boundaries in ultrasound images severely hinder reliable feature extraction and correspondence matching. Existing methods often suffer from poor anatomical alignment and lack physical interpretability. To address the problem, we propose PADReg, a physics-aware deformable registration framework guided by contact force. PADReg leverages synchronized contact force measured by robotic ultrasound systems as a physical prior to constrain the registration. Specifically, instead of directly predicting deformation fields, we first construct a pixel-wise stiffness map utilizing the multi-modal information from contact force and ultrasound images. The stiffness map is then combined with force data to estimate a dense deformation field, through a lightweight physics-aware module inspired by Hooke's law. This design enables PADReg to achieve physically plausible registration with better anatomical alignment than previous methods relying solely on image similarity. Experiments on in-vivo datasets demonstrate that it attains a HD95 of 12.90, which is 21.34\% better than state-of-the-art methods. The source code is available at https://github.com/evelynskip/PADReg.
MM2CT: MR-to-CT translation for multi-modal image fusion with mamba
Aug 07, 2025Abstract:Magnetic resonance (MR)-to-computed tomography (CT) translation offers significant advantages, including the elimination of radiation exposure associated with CT scans and the mitigation of imaging artifacts caused by patient motion. The existing approaches are based on single-modality MR-to-CT translation, with limited research exploring multimodal fusion. To address this limitation, we introduce Multi-modal MR to CT (MM2CT) translation method by leveraging multimodal T1- and T2-weighted MRI data, an innovative Mamba-based framework for multi-modal medical image synthesis. Mamba effectively overcomes the limited local receptive field in CNNs and the high computational complexity issues in Transformers. MM2CT leverages this advantage to maintain long-range dependencies modeling capabilities while achieving multi-modal MR feature integration. Additionally, we incorporate a dynamic local convolution module and a dynamic enhancement module to improve MRI-to-CT synthesis. The experiments on a public pelvis dataset demonstrate that MM2CT achieves state-of-the-art performance in terms of Structural Similarity Index Measure (SSIM) and Peak Signal-to-Noise Ratio (PSNR). Our code is publicly available at https://github.com/Gots-ch/MM2CT.
F2PASeg: Feature Fusion for Pituitary Anatomy Segmentation in Endoscopic Surgery
Aug 07, 2025Abstract:Pituitary tumors often cause deformation or encapsulation of adjacent vital structures. Anatomical structure segmentation can provide surgeons with early warnings of regions that pose surgical risks, thereby enhancing the safety of pituitary surgery. However, pixel-level annotated video stream datasets for pituitary surgeries are extremely rare. To address this challenge, we introduce a new dataset for Pituitary Anatomy Segmentation (PAS). PAS comprises 7,845 time-coherent images extracted from 120 videos. To mitigate class imbalance, we apply data augmentation techniques that simulate the presence of surgical instruments in the training data. One major challenge in pituitary anatomy segmentation is the inconsistency in feature representation due to occlusions, camera motion, and surgical bleeding. By incorporating a Feature Fusion module, F2PASeg is proposed to refine anatomical structure segmentation by leveraging both high-resolution image features and deep semantic embeddings, enhancing robustness against intraoperative variations. Experimental results demonstrate that F2PASeg consistently segments critical anatomical structures in real time, providing a reliable solution for intraoperative pituitary surgery planning. Code: https://github.com/paulili08/F2PASeg.
The Docking Game: Loop Self-Play for Fast, Dynamic, and Accurate Prediction of Flexible Protein--Ligand Binding
Aug 07, 2025Abstract:Molecular docking is a crucial aspect of drug discovery, as it predicts the binding interactions between small-molecule ligands and protein pockets. However, current multi-task learning models for docking often show inferior performance in ligand docking compared to protein pocket docking. This disparity arises largely due to the distinct structural complexities of ligands and proteins. To address this issue, we propose a novel game-theoretic framework that models the protein-ligand interaction as a two-player game called the Docking Game, with the ligand docking module acting as the ligand player and the protein pocket docking module as the protein player. To solve this game, we develop a novel Loop Self-Play (LoopPlay) algorithm, which alternately trains these players through a two-level loop. In the outer loop, the players exchange predicted poses, allowing each to incorporate the other's structural predictions, which fosters mutual adaptation over multiple iterations. In the inner loop, each player dynamically refines its predictions by incorporating its own predicted ligand or pocket poses back into its model. We theoretically show the convergence of LoopPlay, ensuring stable optimization. Extensive experiments conducted on public benchmark datasets demonstrate that LoopPlay achieves approximately a 10\% improvement in predicting accurate binding modes compared to previous state-of-the-art methods. This highlights its potential to enhance the accuracy of molecular docking in drug discovery.
MLLM-CL: Continual Learning for Multimodal Large Language Models
Jun 05, 2025



Abstract:Recent Multimodal Large Language Models (MLLMs) excel in vision-language understanding but face challenges in adapting to dynamic real-world scenarios that require continuous integration of new knowledge and skills. While continual learning (CL) offers a potential solution, existing benchmarks and methods suffer from critical limitations. In this paper, we introduce MLLM-CL, a novel benchmark encompassing domain and ability continual learning, where the former focuses on independently and identically distributed (IID) evaluation across evolving mainstream domains, whereas the latter evaluates on non-IID scenarios with emerging model ability. Methodologically, we propose preventing catastrophic interference through parameter isolation, along with an MLLM-based routing mechanism. Extensive experiments demonstrate that our approach can integrate domain-specific knowledge and functional abilities with minimal forgetting, significantly outperforming existing methods.
Semi-parametric Memory Consolidation: Towards Brain-like Deep Continual Learning
Apr 20, 2025Abstract:Humans and most animals inherently possess a distinctive capacity to continually acquire novel experiences and accumulate worldly knowledge over time. This ability, termed continual learning, is also critical for deep neural networks (DNNs) to adapt to the dynamically evolving world in open environments. However, DNNs notoriously suffer from catastrophic forgetting of previously learned knowledge when trained on sequential tasks. In this work, inspired by the interactive human memory and learning system, we propose a novel biomimetic continual learning framework that integrates semi-parametric memory and the wake-sleep consolidation mechanism. For the first time, our method enables deep neural networks to retain high performance on novel tasks while maintaining prior knowledge in real-world challenging continual learning scenarios, e.g., class-incremental learning on ImageNet. This study demonstrates that emulating biological intelligence provides a promising path to enable deep neural networks with continual learning capabilities.
Pareto Continual Learning: Preference-Conditioned Learning and Adaption for Dynamic Stability-Plasticity Trade-off
Mar 30, 2025Abstract:Continual learning aims to learn multiple tasks sequentially. A key challenge in continual learning is balancing between two objectives: retaining knowledge from old tasks (stability) and adapting to new tasks (plasticity). Experience replay methods, which store and replay past data alongside new data, have become a widely adopted approach to mitigate catastrophic forgetting. However, these methods neglect the dynamic nature of the stability-plasticity trade-off and aim to find a fixed and unchanging balance, resulting in suboptimal adaptation during training and inference. In this paper, we propose Pareto Continual Learning (ParetoCL), a novel framework that reformulates the stability-plasticity trade-off in continual learning as a multi-objective optimization (MOO) problem. ParetoCL introduces a preference-conditioned model to efficiently learn a set of Pareto optimal solutions representing different trade-offs and enables dynamic adaptation during inference. From a generalization perspective, ParetoCL can be seen as an objective augmentation approach that learns from different objective combinations of stability and plasticity. Extensive experiments across multiple datasets and settings demonstrate that ParetoCL outperforms state-of-the-art methods and adapts to diverse continual learning scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge