Dongxia Wu
MF-LAL: Drug Compound Generation Using Multi-Fidelity Latent Space Active Learning
Oct 15, 2024Abstract:Current generative models for drug discovery primarily use molecular docking as an oracle to guide the generation of active compounds. However, such models are often not useful in practice because even compounds with high docking scores do not consistently show experimental activity. More accurate methods for activity prediction exist, such as molecular dynamics based binding free energy calculations, but they are too computationally expensive to use in a generative model. To address this challenge, we propose Multi-Fidelity Latent space Active Learning (MF-LAL), a generative modeling framework that integrates a set of oracles with varying cost-accuracy tradeoffs. Unlike previous approaches that separately learn the surrogate model and generative model, MF-LAL combines the generative and multi-fidelity surrogate models into a single framework, allowing for more accurate activity prediction and higher quality samples. We train MF-LAL with a novel active learning algorithm to further reduce computational cost. Our experiments on two disease-relevant proteins show that MF-LAL produces compounds with significantly better binding free energy scores than other single and multi-fidelity approaches.
Functional-level Uncertainty Quantification for Calibrated Fine-tuning on LLMs
Oct 09, 2024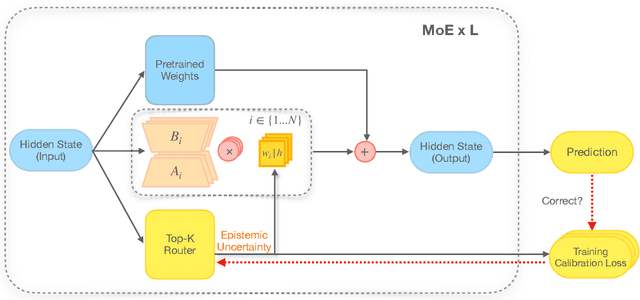
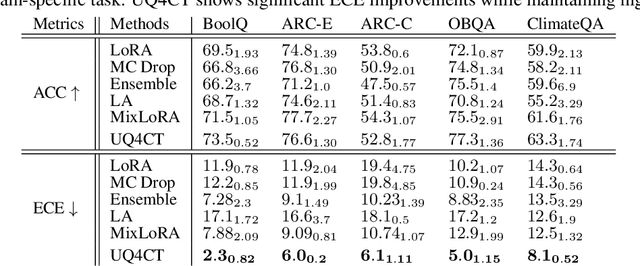
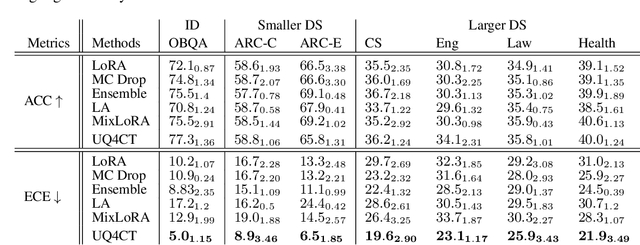
Abstract:From common-sense reasoning to domain-specific tasks, parameter-efficient fine tuning (PEFT) methods for large language models (LLMs) have showcased significant performance improvements on downstream tasks. However, fine-tuned LLMs often struggle with overconfidence in uncertain predictions, particularly due to sparse training data. This overconfidence reflects poor epistemic uncertainty calibration, which arises from limitations in the model's ability to generalize with limited data. Existing PEFT uncertainty quantification methods for LLMs focus on the post fine-tuning stage and thus have limited capability in calibrating epistemic uncertainty. To address these limitations, we propose Functional-Level Uncertainty Quantification for Calibrated Fine-Tuning (UQ4CT), which captures and calibrates functional-level epistemic uncertainty during the fine-tuning stage via a mixture-of-expert framework. We show that UQ4CT reduces Expected Calibration Error (ECE) by more than $25\%$ while maintaining high accuracy across $5$ benchmarks. Furthermore, UQ4CT maintains superior ECE performance with high accuracy under distribution shift, showcasing improved generalizability.
Diff-BBO: Diffusion-Based Inverse Modeling for Black-Box Optimization
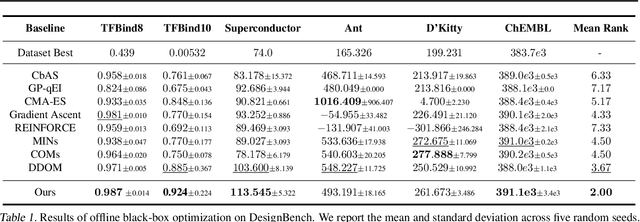
Jun 30, 2024Abstract:Black-box optimization (BBO) aims to optimize an objective function by iteratively querying a black-box oracle. This process demands sample-efficient optimization due to the high computational cost of function evaluations. While prior studies focus on forward approaches to learn surrogates for the unknown objective function, they struggle with high-dimensional inputs where valid inputs form a small subspace (e.g., valid protein sequences), which is common in real-world tasks. Recently, diffusion models have demonstrated impressive capability in learning the high-dimensional data manifold. They have shown promising performance in black-box optimization tasks but only in offline settings. In this work, we propose diffusion-based inverse modeling for black-box optimization (Diff-BBO), the first inverse approach leveraging diffusion models for online BBO problem. Diff-BBO distinguishes itself from forward approaches through the design of acquisition function. Instead of proposing candidates in the design space, Diff-BBO employs a novel acquisition function Uncertainty-aware Exploration (UaE) to propose objective function values, which leverages the uncertainty of a conditional diffusion model to generate samples in the design space. Theoretically, we prove that using UaE leads to optimal optimization outcomes. Empirically, we redesign experiments on the Design-Bench benchmark for online settings and show that Diff-BBO achieves state-of-the-art performance.
Multi-Fidelity Residual Neural Processes for Scalable Surrogate Modeling
Feb 29, 2024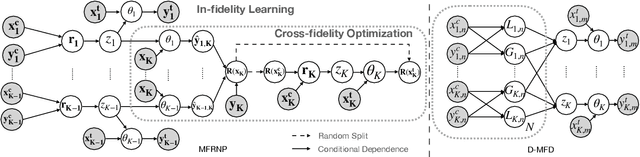
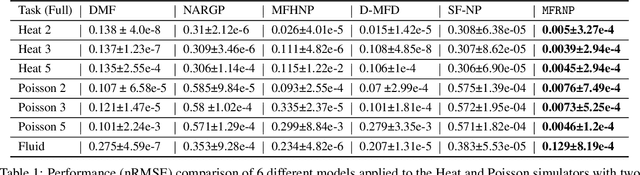
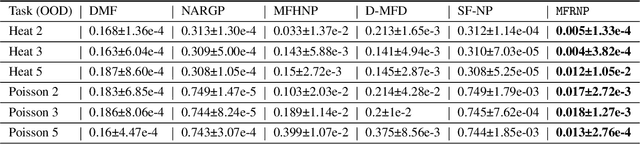
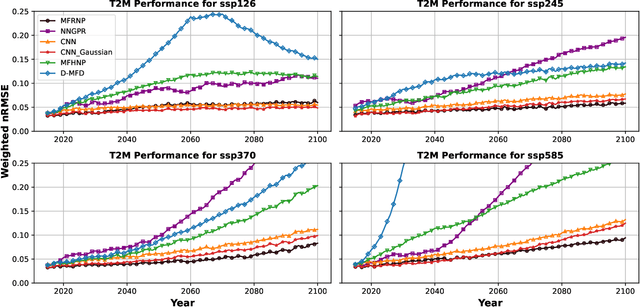
Abstract:Multi-fidelity surrogate modeling aims to learn an accurate surrogate at the highest fidelity level by combining data from multiple sources. Traditional methods relying on Gaussian processes can hardly scale to high-dimensional data. Deep learning approaches utilize neural network based encoders and decoders to improve scalability. These approaches share encoded representations across fidelities without including corresponding decoder parameters. At the highest fidelity, the representations are decoded with different parameters, making the shared information inherently inaccurate. This hinders inference performance, especially in out-of-distribution scenarios when the highest fidelity data has limited domain coverage. To address these limitations, we propose Multi-fidelity Residual Neural Processes (MFRNP), a novel multi-fidelity surrogate modeling framework. MFRNP optimizes lower fidelity decoders for accurate information sharing by aggregating lower fidelity surrogate outputs and models residual between the aggregation and ground truth on the highest fidelity. We show that MFRNP significantly outperforms current state-of-the-art in learning partial differential equations and a real-world climate modeling task.
Diffusion Models as Constrained Samplers for Optimization with Unknown Constraints
Feb 28, 2024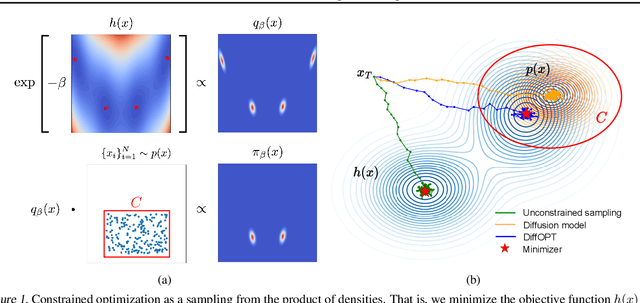
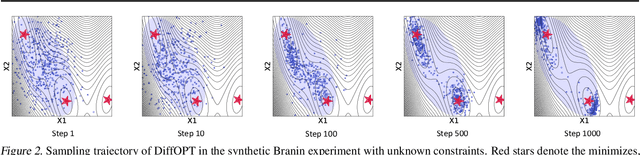
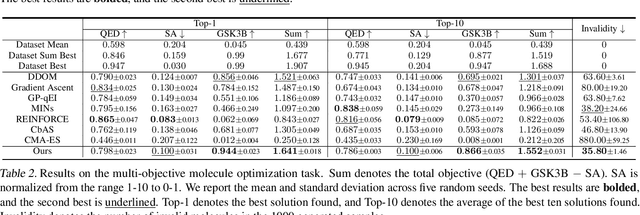
Abstract:Addressing real-world optimization problems becomes particularly challenging when analytic objective functions or constraints are unavailable. While numerous studies have addressed the issue of unknown objectives, limited research has focused on scenarios where feasibility constraints are not given explicitly. Overlooking these constraints can lead to spurious solutions that are unrealistic in practice. To deal with such unknown constraints, we propose to perform optimization within the data manifold using diffusion models. To constrain the optimization process to the data manifold, we reformulate the original optimization problem as a sampling problem from the product of the Boltzmann distribution defined by the objective function and the data distribution learned by the diffusion model. To enhance sampling efficiency, we propose a two-stage framework that begins with a guided diffusion process for warm-up, followed by a Langevin dynamics stage for further correction. Theoretical analysis shows that the initial stage results in a distribution focused on feasible solutions, thereby providing a better initialization for the later stage. Comprehensive experiments on a synthetic dataset, six real-world black-box optimization datasets, and a multi-objective optimization dataset show that our method achieves better or comparable performance with previous state-of-the-art baselines.
MFBind: a Multi-Fidelity Approach for Evaluating Drug Compounds in Practical Generative Modeling
Feb 16, 2024Abstract:Current generative models for drug discovery primarily use molecular docking to evaluate the quality of generated compounds. However, such models are often not useful in practice because even compounds with high docking scores do not consistently show experimental activity. More accurate methods for activity prediction exist, such as molecular dynamics based binding free energy calculations, but they are too computationally expensive to use in a generative model. We propose a multi-fidelity approach, Multi-Fidelity Bind (MFBind), to achieve the optimal trade-off between accuracy and computational cost. MFBind integrates docking and binding free energy simulators to train a multi-fidelity deep surrogate model with active learning. Our deep surrogate model utilizes a pretraining technique and linear prediction heads to efficiently fit small amounts of high-fidelity data. We perform extensive experiments and show that MFBind (1) outperforms other state-of-the-art single and multi-fidelity baselines in surrogate modeling, and (2) boosts the performance of generative models with markedly higher quality compounds.
Learning Granger Causality from Instance-wise Self-attentive Hawkes Processes
Feb 06, 2024Abstract:We address the problem of learning Granger causality from asynchronous, interdependent, multi-type event sequences. In particular, we are interested in discovering instance-level causal structures in an unsupervised manner. Instance-level causality identifies causal relationships among individual events, providing more fine-grained information for decision-making. Existing work in the literature either requires strong assumptions, such as linearity in the intensity function, or heuristically defined model parameters that do not necessarily meet the requirements of Granger causality. We propose Instance-wise Self-Attentive Hawkes Processes (ISAHP), a novel deep learning framework that can directly infer the Granger causality at the event instance level. ISAHP is the first neural point process model that meets the requirements of Granger causality. It leverages the self-attention mechanism of the transformer to align with the principles of Granger causality. We empirically demonstrate that ISAHP is capable of discovering complex instance-level causal structures that cannot be handled by classical models. We also show that ISAHP achieves state-of-the-art performance in proxy tasks involving type-level causal discovery and instance-level event type prediction.
Disentangled Multi-Fidelity Deep Bayesian Active Learning
May 07, 2023Abstract:To balance quality and cost, various domain areas of science and engineering run simulations at multiple levels of sophistication. Multi-fidelity active learning aims to learn a direct mapping from input parameters to simulation outputs by actively acquiring data from multiple fidelity levels. However, existing approaches based on Gaussian processes are hardly scalable to high-dimensional data. Other deep learning-based methods use the hierarchical structure, which only supports passing information from low-fidelity to high-fidelity. This approach also leads to the undesirable propagation of errors from low-fidelity representations to high-fidelity ones. We propose a novel disentangled deep Bayesian learning framework for multi-fidelity active learning, that learns the surrogate models conditioned on the distribution of functions at multiple fidelities.
Multi-fidelity Hierarchical Neural Processes
Jun 10, 2022



Abstract:Science and engineering fields use computer simulation extensively. These simulations are often run at multiple levels of sophistication to balance accuracy and efficiency. Multi-fidelity surrogate modeling reduces the computational cost by fusing different simulation outputs. Cheap data generated from low-fidelity simulators can be combined with limited high-quality data generated by an expensive high-fidelity simulator. Existing methods based on Gaussian processes rely on strong assumptions of the kernel functions and can hardly scale to high-dimensional settings. We propose Multi-fidelity Hierarchical Neural Processes (MF-HNP), a unified neural latent variable model for multi-fidelity surrogate modeling. MF-HNP inherits the flexibility and scalability of Neural Processes. The latent variables transform the correlations among different fidelity levels from observations to latent space. The predictions across fidelities are conditionally independent given the latent states. It helps alleviate the error propagation issue in existing methods. MF-HNP is flexible enough to handle non-nested high dimensional data at different fidelity levels with varying input and output dimensions. We evaluate MF-HNP on epidemiology and climate modeling tasks, achieving competitive performance in terms of accuracy and uncertainty estimation. In contrast to deep Gaussian Processes with only low-dimensional (< 10) tasks, our method shows great promise for speeding up high-dimensional complex simulations (over 7000 for epidemiology modeling and 45000 for climate modeling).
Multi defect detection and analysis of electron microscopy images with deep learning
Aug 19, 2021



Abstract:Electron microscopy is widely used to explore defects in crystal structures, but human detecting of defects is often time-consuming, error-prone, and unreliable, and is not scalable to large numbers of images or real-time analysis. In this work, we discuss the application of machine learning approaches to find the location and geometry of different defect clusters in irradiated steels. We show that a deep learning based Faster R-CNN analysis system has a performance comparable to human analysis with relatively small training data sets. This study proves the promising ability to apply deep learning to assist the development of automated microscopy data analysis even when multiple features are present and paves the way for fast, scalable, and reliable analysis systems for massive amounts of modern electron microscopy data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge