Matteo Chinazzi
Disentangled Multi-Fidelity Deep Bayesian Active Learning
May 07, 2023Abstract:To balance quality and cost, various domain areas of science and engineering run simulations at multiple levels of sophistication. Multi-fidelity active learning aims to learn a direct mapping from input parameters to simulation outputs by actively acquiring data from multiple fidelity levels. However, existing approaches based on Gaussian processes are hardly scalable to high-dimensional data. Other deep learning-based methods use the hierarchical structure, which only supports passing information from low-fidelity to high-fidelity. This approach also leads to the undesirable propagation of errors from low-fidelity representations to high-fidelity ones. We propose a novel disentangled deep Bayesian learning framework for multi-fidelity active learning, that learns the surrogate models conditioned on the distribution of functions at multiple fidelities.
Multi-fidelity Hierarchical Neural Processes
Jun 10, 2022



Abstract:Science and engineering fields use computer simulation extensively. These simulations are often run at multiple levels of sophistication to balance accuracy and efficiency. Multi-fidelity surrogate modeling reduces the computational cost by fusing different simulation outputs. Cheap data generated from low-fidelity simulators can be combined with limited high-quality data generated by an expensive high-fidelity simulator. Existing methods based on Gaussian processes rely on strong assumptions of the kernel functions and can hardly scale to high-dimensional settings. We propose Multi-fidelity Hierarchical Neural Processes (MF-HNP), a unified neural latent variable model for multi-fidelity surrogate modeling. MF-HNP inherits the flexibility and scalability of Neural Processes. The latent variables transform the correlations among different fidelity levels from observations to latent space. The predictions across fidelities are conditionally independent given the latent states. It helps alleviate the error propagation issue in existing methods. MF-HNP is flexible enough to handle non-nested high dimensional data at different fidelity levels with varying input and output dimensions. We evaluate MF-HNP on epidemiology and climate modeling tasks, achieving competitive performance in terms of accuracy and uncertainty estimation. In contrast to deep Gaussian Processes with only low-dimensional (< 10) tasks, our method shows great promise for speeding up high-dimensional complex simulations (over 7000 for epidemiology modeling and 45000 for climate modeling).
Accelerating Stochastic Simulation with Interactive Neural Processes
Jun 11, 2021



Abstract:Stochastic simulations such as large-scale, spatiotemporal, age-structured epidemic models are computationally expensive at fine-grained resolution. We propose Interactive Neural Process (INP), an interactive framework to continuously learn a deep learning surrogate model and accelerate simulation. Our framework is based on the novel integration of Bayesian active learning, stochastic simulation and deep sequence modeling. In particular, we develop a novel spatiotemporal neural process model to mimic the underlying process dynamics. Our model automatically infers the latent process which describes the intrinsic uncertainty of the simulator. This also gives rise to a new acquisition function that can quantify the uncertainty of deep learning predictions. We design Bayesian active learning algorithms to iteratively query the simulator, gather more data, and continuously improve the model. We perform theoretical analysis and demonstrate that our approach reduces sample complexity compared with random sampling in high dimension. Empirically, we demonstrate our framework can faithfully imitate the behavior of a complex infectious disease simulator with a small number of examples, enabling rapid simulation and scenario exploration.
Quantifying Uncertainty in Deep Spatiotemporal Forecasting
May 25, 2021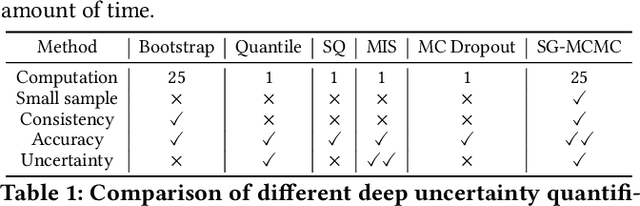
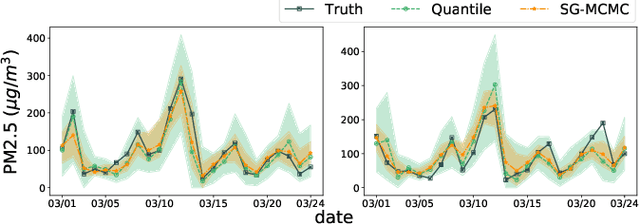
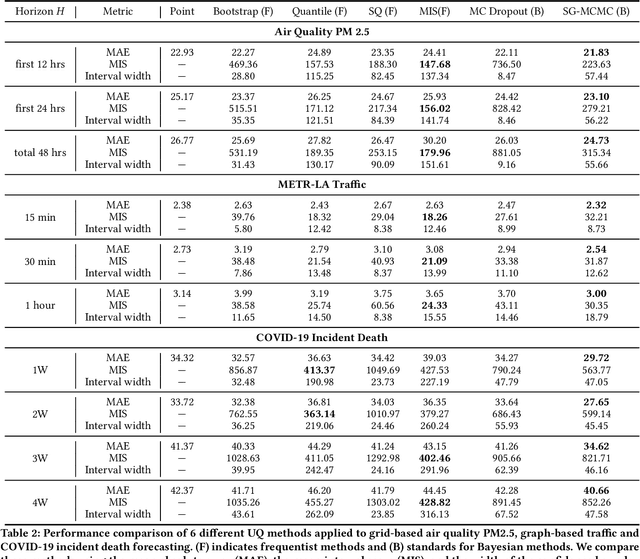
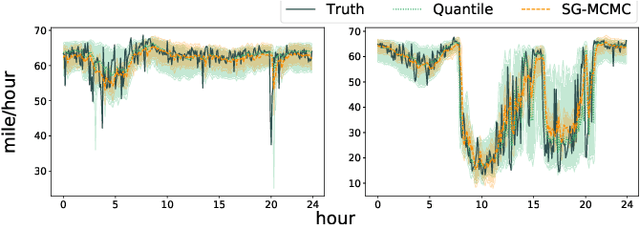
Abstract:Deep learning is gaining increasing popularity for spatiotemporal forecasting. However, prior works have mostly focused on point estimates without quantifying the uncertainty of the predictions. In high stakes domains, being able to generate probabilistic forecasts with confidence intervals is critical to risk assessment and decision making. Hence, a systematic study of uncertainty quantification (UQ) methods for spatiotemporal forecasting is missing in the community. In this paper, we describe two types of spatiotemporal forecasting problems: regular grid-based and graph-based. Then we analyze UQ methods from both the Bayesian and the frequentist point of view, casting in a unified framework via statistical decision theory. Through extensive experiments on real-world road network traffic, epidemics, and air quality forecasting tasks, we reveal the statistical and computational trade-offs for different UQ methods: Bayesian methods are typically more robust in mean prediction, while confidence levels obtained from frequentist methods provide more extensive coverage over data variations. Computationally, quantile regression type methods are cheaper for a single confidence interval but require re-training for different intervals. Sampling based methods generate samples that can form multiple confidence intervals, albeit at a higher computational cost.
DeepGLEAM: a hybrid mechanistic and deep learning model for COVID-19 forecasting
Feb 15, 2021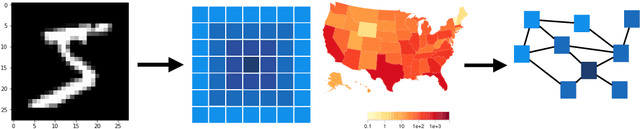
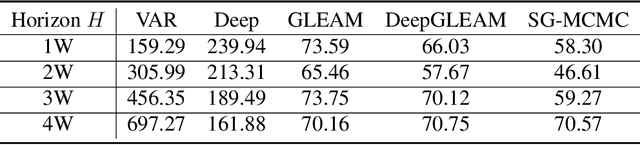
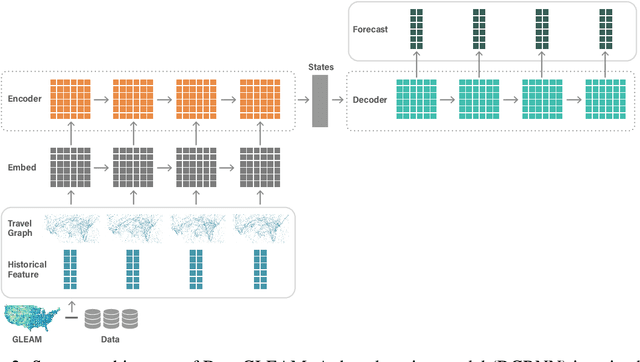
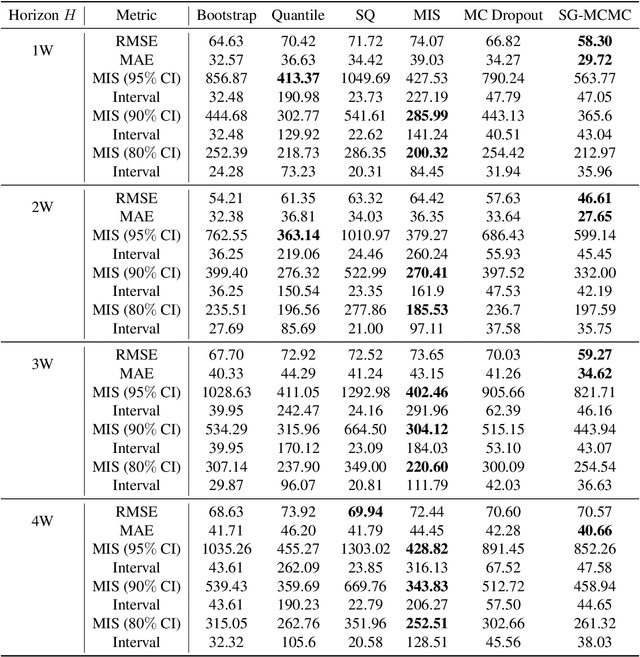
Abstract:We introduce DeepGLEAM, a hybrid model for COVID-19 forecasting. DeepGLEAM combines a mechanistic stochastic simulation model GLEAM with deep learning. It uses deep learning to learn the correction terms from GLEAM, which leads to improved performance. We further integrate various uncertainty quantification methods to generate confidence intervals. We demonstrate DeepGLEAM on real-world COVID-19 mortality forecasting tasks.
Finding Patient Zero: Learning Contagion Source with Graph Neural Networks
Jun 27, 2020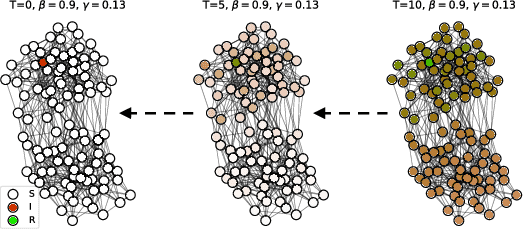
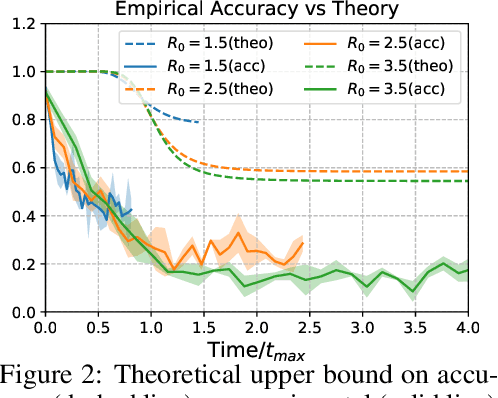
Abstract:Locating the source of an epidemic, or patient zero (P0), can provide critical insights into the infection's transmission course and allow efficient resource allocation. Existing methods use graph-theoretic centrality measures and expensive message-passing algorithms, requiring knowledge of the underlying dynamics and its parameters. In this paper, we revisit this problem using graph neural networks (GNNs) to learn P0. We establish a theoretical limit for the identification of P0 in a class of epidemic models. We evaluate our method against different epidemic models on both synthetic and a real-world contact network considering a disease with history and characteristics of COVID-19. % We observe that GNNs can identify P0 close to the theoretical bound on accuracy, without explicit input of dynamics or its parameters. In addition, GNN is over 100 times faster than classic methods for inference on arbitrary graph topologies. Our theoretical bound also shows that the epidemic is like a ticking clock, emphasizing the importance of early contact-tracing. We find a maximum time after which accurate recovery of the source becomes impossible, regardless of the algorithm used.
A machine learning methodology for real-time forecasting of the 2019-2020 COVID-19 outbreak using Internet searches, news alerts, and estimates from mechanistic models
Apr 08, 2020



Abstract:We present a timely and novel methodology that combines disease estimates from mechanistic models with digital traces, via interpretable machine-learning methodologies, to reliably forecast COVID-19 activity in Chinese provinces in real-time. Specifically, our method is able to produce stable and accurate forecasts 2 days ahead of current time, and uses as inputs (a) official health reports from Chinese Center Disease for Control and Prevention (China CDC), (b) COVID-19-related internet search activity from Baidu, (c) news media activity reported by Media Cloud, and (d) daily forecasts of COVID-19 activity from GLEAM, an agent-based mechanistic model. Our machine-learning methodology uses a clustering technique that enables the exploitation of geo-spatial synchronicities of COVID-19 activity across Chinese provinces, and a data augmentation technique to deal with the small number of historical disease activity observations, characteristic of emerging outbreaks. Our model's predictive power outperforms a collection of baseline models in 27 out of the 32 Chinese provinces, and could be easily extended to other geographies currently affected by the COVID-19 outbreak to help decision makers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge