Nathaniel J. Krakauer
Multi defect detection and analysis of electron microscopy images with deep learning
Aug 19, 2021



Abstract:Electron microscopy is widely used to explore defects in crystal structures, but human detecting of defects is often time-consuming, error-prone, and unreliable, and is not scalable to large numbers of images or real-time analysis. In this work, we discuss the application of machine learning approaches to find the location and geometry of different defect clusters in irradiated steels. We show that a deep learning based Faster R-CNN analysis system has a performance comparable to human analysis with relatively small training data sets. This study proves the promising ability to apply deep learning to assist the development of automated microscopy data analysis even when multiple features are present and paves the way for fast, scalable, and reliable analysis systems for massive amounts of modern electron microscopy data.
Assessing Graph-based Deep Learning Models for Predicting Flash Point
Feb 26, 2020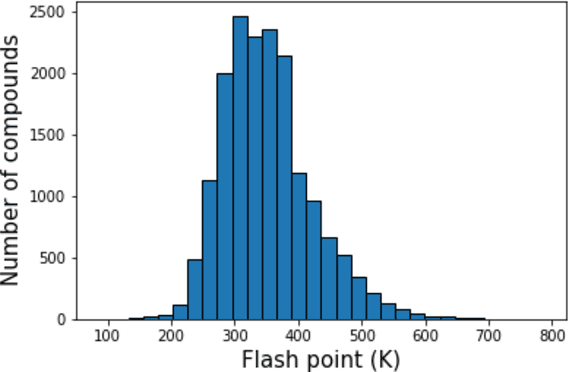
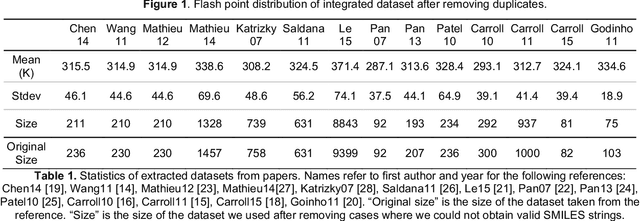
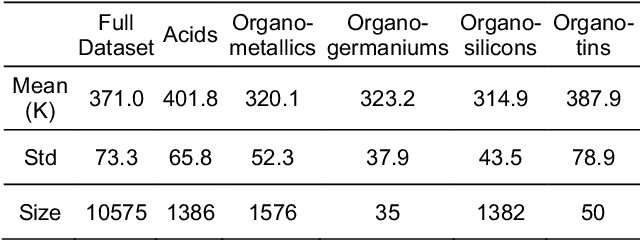
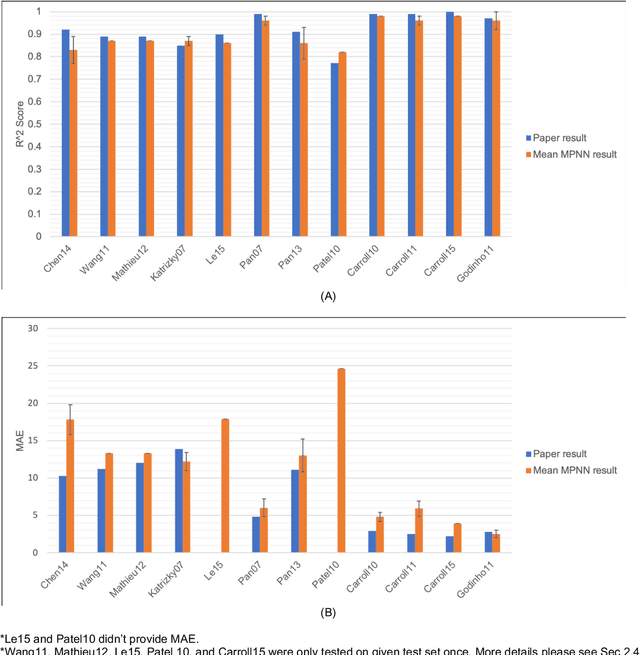
Abstract:Flash points of organic molecules play an important role in preventing flammability hazards and large databases of measured values exist, although millions of compounds remain unmeasured. To rapidly extend existing data to new compounds many researchers have used quantitative structure-property relationship (QSPR) analysis to effectively predict flash points. In recent years graph-based deep learning (GBDL) has emerged as a powerful alternative method to traditional QSPR. In this paper, GBDL models were implemented in predicting flash point for the first time. We assessed the performance of two GBDL models, message-passing neural network (MPNN) and graph convolutional neural network (GCNN), by comparing methods. Our result shows that MPNN both outperforms GCNN and yields slightly worse but comparable performance with previous QSPR studies. The average R2 and Mean Absolute Error (MAE) scores of MPNN are, respectively, 2.3% lower and 2.0 K higher than previous comparable studies. To further explore GBDL models, we collected the largest flash point dataset to date, which contains 10575 unique molecules. The optimized MPNN gives a test data R2 of 0.803 and MAE of 17.8 K on the complete dataset. We also extracted 5 datasets from our integrated dataset based on molecular types (acids, organometallics, organogermaniums, organosilicons, and organotins) and explore the quality of the model in these classes.against 12 previous QSPR studies using more traditional
* 26 pages, 6 tabels, 3 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge