David Belanger
AT&T Inc, Stevens Institute of Technology
The Next Decade of Telecommunications Artificial Intelligence
Feb 22, 2021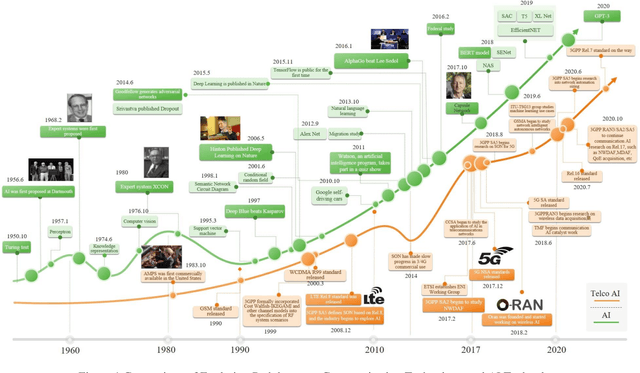
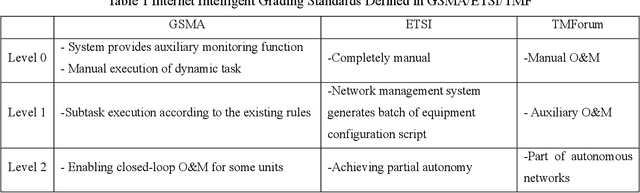
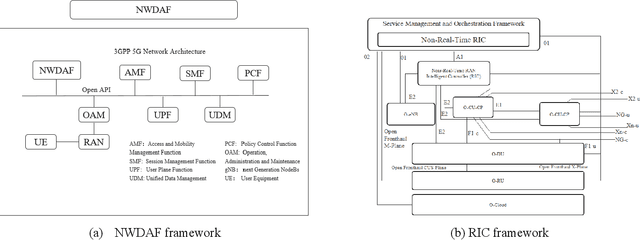
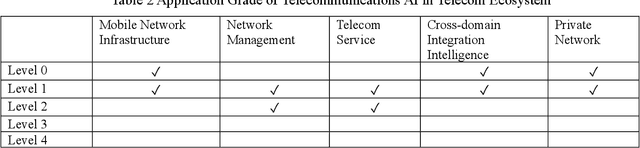
Abstract:It has been an exciting journey since the mobile communications and artificial intelligence were conceived 37 years and 64 years ago. While both fields evolved independently and profoundly changed communications and computing industries, the rapid convergence of 5G and deep learning is beginning to significantly transform the core communication infrastructure, network management and vertical applications. The paper first outlines the individual roadmaps of mobile communications and artificial intelligence in the early stage, with a concentration to review the era from 3G to 5G when AI and mobile communications started to converge. With regard to telecommunications artificial intelligence, the paper further introduces in detail the progress of artificial intelligence in the ecosystem of mobile communications. The paper then summarizes the classifications of AI in telecom ecosystems along with its evolution paths specified by various international telecommunications standardization bodies. Towards the next decade, the paper forecasts the prospective roadmap of telecommunications artificial intelligence. In line with 3GPP and ITU-R timeline of 5G & 6G, the paper further explores the network intelligence following 3GPP and ORAN routes respectively, experience and intention driven network management and operation, network AI signalling system, intelligent middle-office based BSS, intelligent customer experience management and policy control driven by BSS and OSS convergence, evolution from SLA to ELA, and intelligent private network for verticals. The paper is concluded with the vision that AI will reshape the future B5G or 6G landscape and we need pivot our R&D, standardizations, and ecosystem to fully take the unprecedented opportunities.
Is Transfer Learning Necessary for Protein Landscape Prediction?
Oct 31, 2020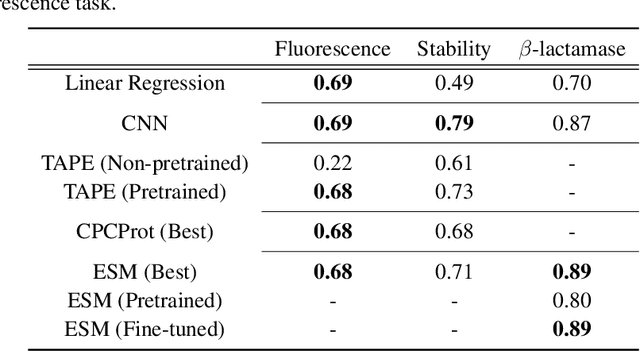
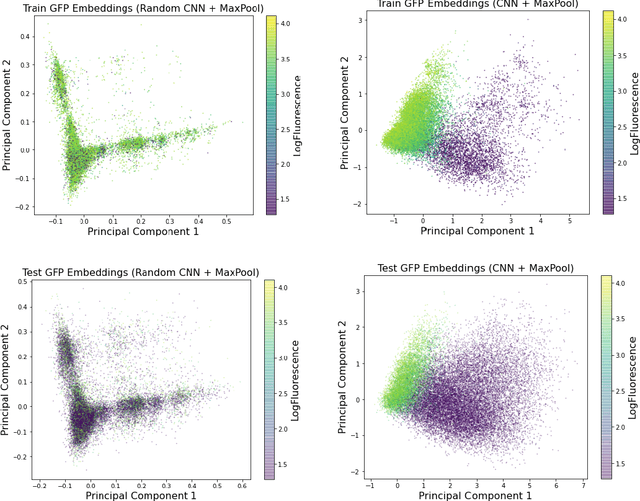
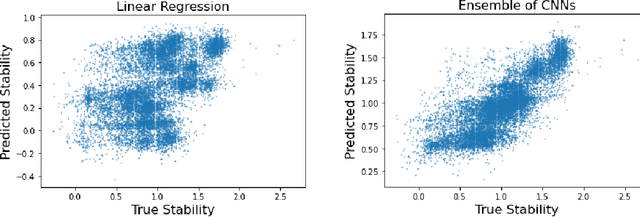
Abstract:Recently, there has been great interest in learning how to best represent proteins, specifically with fixed-length embeddings. Deep learning has become a popular tool for protein representation learning as a model's hidden layers produce potentially useful vector embeddings. TAPE introduced a number of benchmark tasks and showed that semi-supervised learning, via pretraining language models on a large protein corpus, improved performance on downstream tasks. Two of the tasks (fluorescence prediction and stability prediction) involve learning fitness landscapes. In this paper, we show that CNN models trained solely using supervised learning both compete with and sometimes outperform the best models from TAPE that leverage expensive pretraining on large protein datasets. These CNN models are sufficiently simple and small that they can be trained using a Google Colab notebook. We also find for the fluorescence task that linear regression outperforms our models and the TAPE models. The benchmarking tasks proposed by TAPE are excellent measures of a model's ability to predict protein function and should be used going forward. However, we believe it is important to add baselines from simple models to put the performance of the semi-supervised models that have been reported so far into perspective.
Fixed-Length Protein Embeddings using Contextual Lenses
Oct 15, 2020


Abstract:The Basic Local Alignment Search Tool (BLAST) is currently the most popular method for searching databases of biological sequences. BLAST compares sequences via similarity defined by a weighted edit distance, which results in it being computationally expensive. As opposed to working with edit distance, a vector similarity approach can be accelerated substantially using modern hardware or hashing techniques. Such an approach would require fixed-length embeddings for biological sequences. There has been recent interest in learning fixed-length protein embeddings using deep learning models under the hypothesis that the hidden layers of supervised or semi-supervised models could produce potentially useful vector embeddings. We consider transformer (BERT) protein language models that are pretrained on the TrEMBL data set and learn fixed-length embeddings on top of them with contextual lenses. The embeddings are trained to predict the family a protein belongs to for sequences in the Pfam database. We show that for nearest-neighbor family classification, pretraining offers a noticeable boost in performance and that the corresponding learned embeddings are competitive with BLAST. Furthermore, we show that the raw transformer embeddings, obtained via static pooling, do not perform well on nearest-neighbor family classification, which suggests that learning embeddings in a supervised manner via contextual lenses may be a compute-efficient alternative to fine-tuning.
Rethinking Attention with Performers
Sep 30, 2020

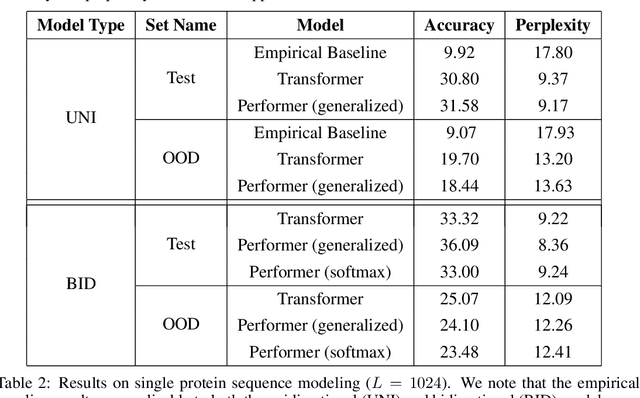
Abstract:We introduce Performers, Transformer architectures which can estimate regular (softmax) full-rank-attention Transformers with provable accuracy, but using only linear (as opposed to quadratic) space and time complexity, without relying on any priors such as sparsity or low-rankness. To approximate softmax attention-kernels, Performers use a novel Fast Attention Via positive Orthogonal Random features approach (FAVOR+), which may be of independent interest for scalable kernel methods. FAVOR+ can be also used to efficiently model kernelizable attention mechanisms beyond softmax. This representational power is crucial to accurately compare softmax with other kernels for the first time on large-scale tasks, beyond the reach of regular Transformers, and investigate optimal attention-kernels. Performers are linear architectures fully compatible with regular Transformers and with strong theoretical guarantees: unbiased or nearly-unbiased estimation of the attention matrix, uniform convergence and low estimation variance. We tested Performers on a rich set of tasks stretching from pixel-prediction through text models to protein sequence modeling. We demonstrate competitive results with other examined efficient sparse and dense attention methods, showcasing effectiveness of the novel attention-learning paradigm leveraged by Performers.
Masked Language Modeling for Proteins via Linearly Scalable Long-Context Transformers
Jun 05, 2020



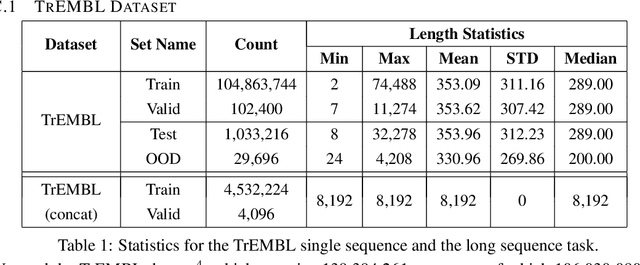
Abstract:Transformer models have achieved state-of-the-art results across a diverse range of domains. However, concern over the cost of training the attention mechanism to learn complex dependencies between distant inputs continues to grow. In response, solutions that exploit the structure and sparsity of the learned attention matrix have blossomed. However, real-world applications that involve long sequences, such as biological sequence analysis, may fall short of meeting these assumptions, precluding exploration of these models. To address this challenge, we present a new Transformer architecture, Performer, based on Fast Attention Via Orthogonal Random features (FAVOR). Our mechanism scales linearly rather than quadratically in the number of tokens in the sequence, is characterized by sub-quadratic space complexity and does not incorporate any sparsity pattern priors. Furthermore, it provides strong theoretical guarantees: unbiased estimation of the attention matrix and uniform convergence. It is also backwards-compatible with pre-trained regular Transformers. We demonstrate its effectiveness on the challenging task of protein sequence modeling and provide detailed theoretical analysis.
Population-Based Black-Box Optimization for Biological Sequence Design
Jun 05, 2020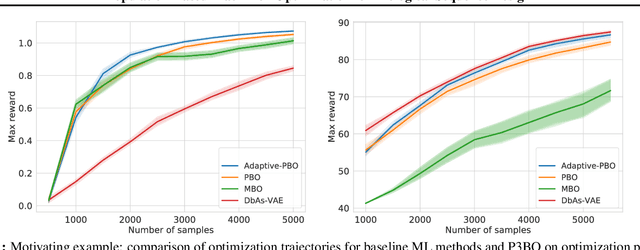
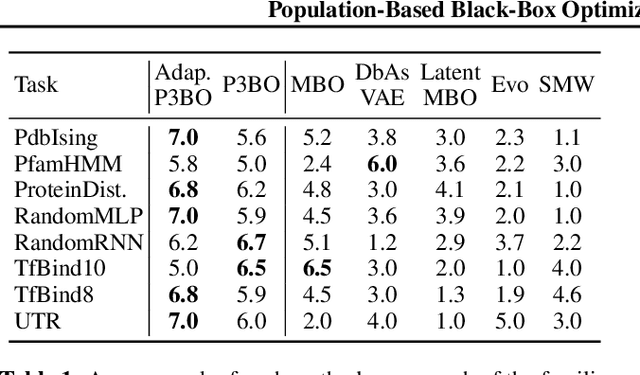
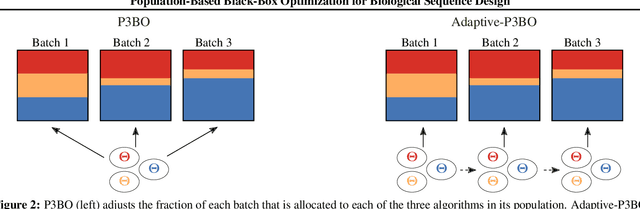
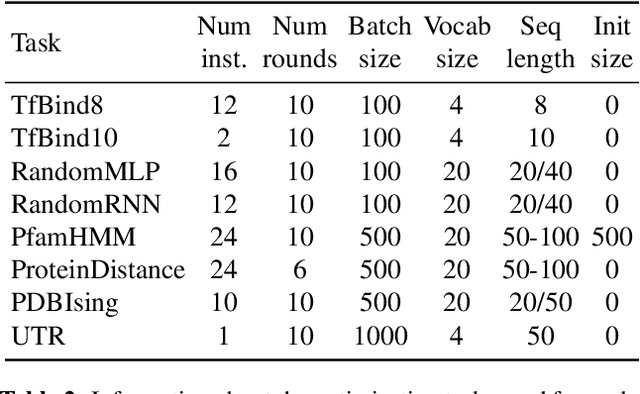
Abstract:The use of black-box optimization for the design of new biological sequences is an emerging research area with potentially revolutionary impact. The cost and latency of wet-lab experiments requires methods that find good sequences in few experimental rounds of large batches of sequences--a setting that off-the-shelf black-box optimization methods are ill-equipped to handle. We find that the performance of existing methods varies drastically across optimization tasks, posing a significant obstacle to real-world applications. To improve robustness, we propose Population-Based Black-Box Optimization (P3BO), which generates batches of sequences by sampling from an ensemble of methods. The number of sequences sampled from any method is proportional to the quality of sequences it previously proposed, allowing P3BO to combine the strengths of individual methods while hedging against their innate brittleness. Adapting the hyper-parameters of each of the methods online using evolutionary optimization further improves performance. Through extensive experiments on in-silico optimization tasks, we show that P3BO outperforms any single method in its population, proposing higher quality sequences as well as more diverse batches. As such, P3BO and Adaptive-P3BO are a crucial step towards deploying ML to real-world sequence design.
Boundless: Generative Adversarial Networks for Image Extension
Aug 19, 2019
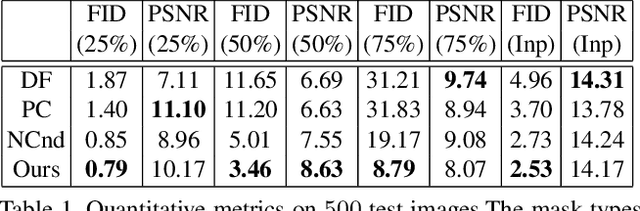
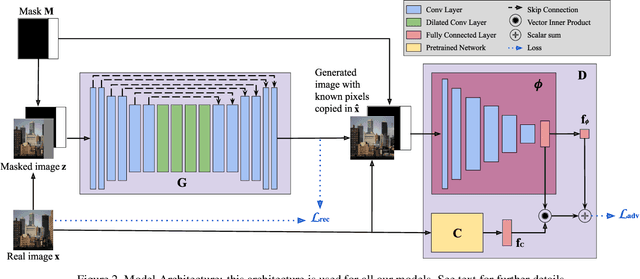
Abstract:Image extension models have broad applications in image editing, computational photography and computer graphics. While image inpainting has been extensively studied in the literature, it is challenging to directly apply the state-of-the-art inpainting methods to image extension as they tend to generate blurry or repetitive pixels with inconsistent semantics. We introduce semantic conditioning to the discriminator of a generative adversarial network (GAN), and achieve strong results on image extension with coherent semantics and visually pleasing colors and textures. We also show promising results in extreme extensions, such as panorama generation.
Predicting Electron-Ionization Mass Spectrometry using Neural Networks
Nov 21, 2018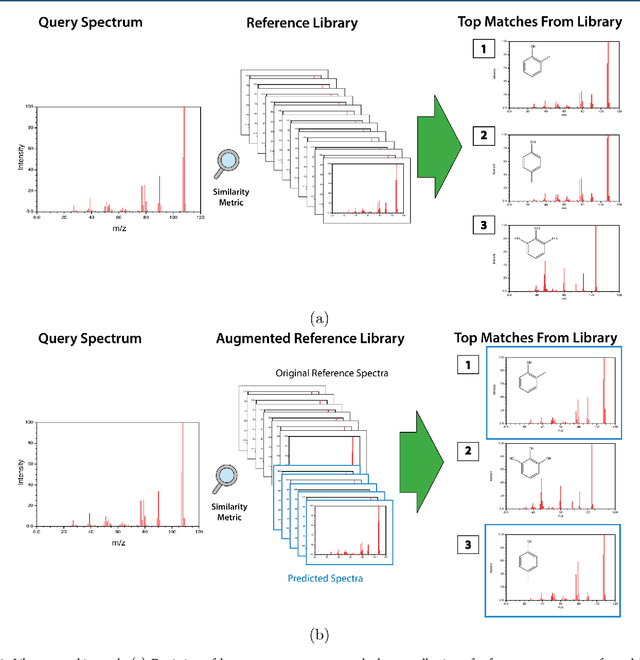
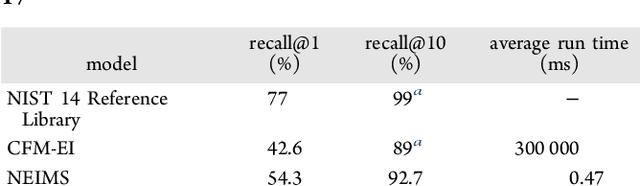
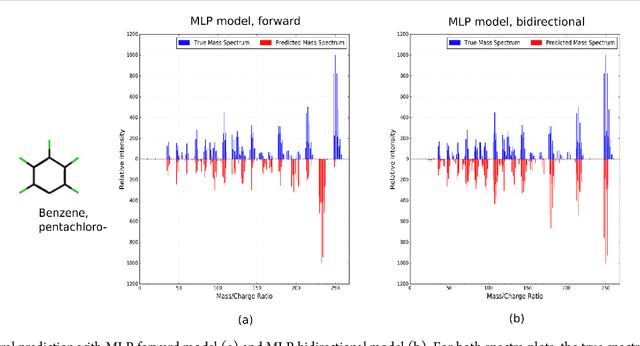
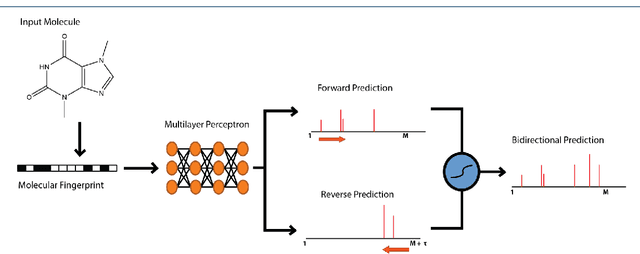
Abstract:When confronted with a substance of unknown identity, researchers often perform mass spectrometry on the sample and compare the observed spectrum to a library of previously-collected spectra to identify the molecule. While popular, this approach will fail to identify molecules that are not in the existing library. In response, we propose to improve the library's coverage by augmenting it with synthetic spectra that are predicted using machine learning. We contribute a lightweight neural network model that quickly predicts mass spectra for small molecules. Achieving high accuracy predictions requires a novel neural network architecture that is designed to capture typical fragmentation patterns from electron ionization. We analyze the effects of our modeling innovations on library matching performance and compare our models to prior machine learning-based work on spectrum prediction.
Learning Latent Permutations with Gumbel-Sinkhorn Networks
Feb 23, 2018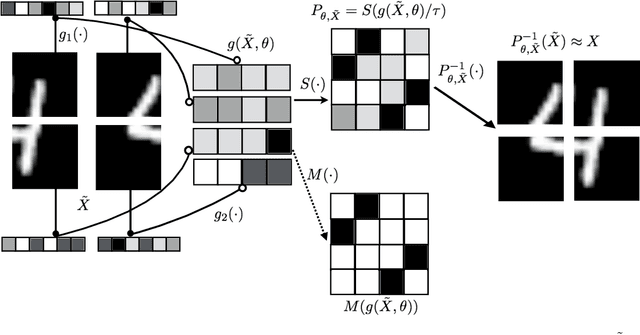



Abstract:Permutations and matchings are core building blocks in a variety of latent variable models, as they allow us to align, canonicalize, and sort data. Learning in such models is difficult, however, because exact marginalization over these combinatorial objects is intractable. In response, this paper introduces a collection of new methods for end-to-end learning in such models that approximate discrete maximum-weight matching using the continuous Sinkhorn operator. Sinkhorn iteration is attractive because it functions as a simple, easy-to-implement analog of the softmax operator. With this, we can define the Gumbel-Sinkhorn method, an extension of the Gumbel-Softmax method (Jang et al. 2016, Maddison2016 et al. 2016) to distributions over latent matchings. We demonstrate the effectiveness of our method by outperforming competitive baselines on a range of qualitatively different tasks: sorting numbers, solving jigsaw puzzles, and identifying neural signals in worms.
Synthesizing Normalized Faces from Facial Identity Features
Oct 17, 2017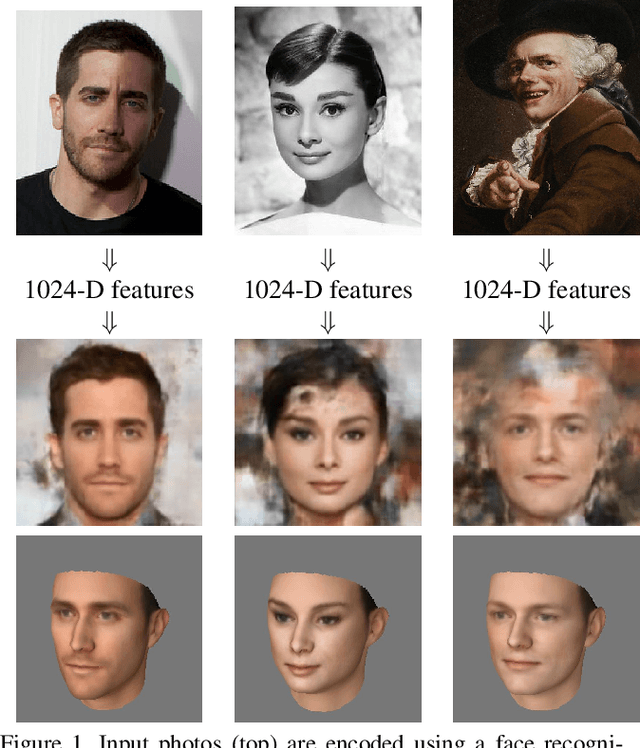
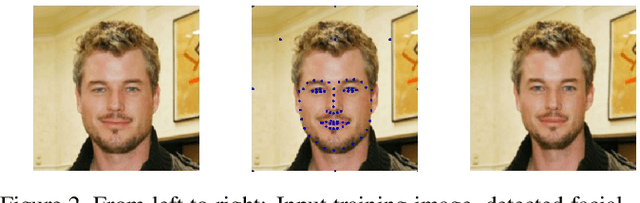
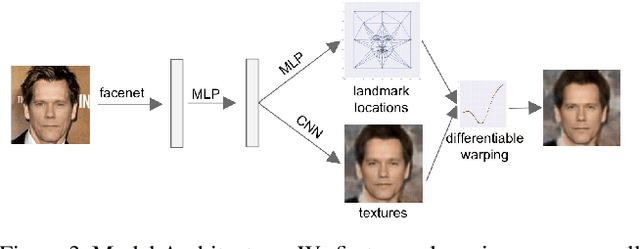
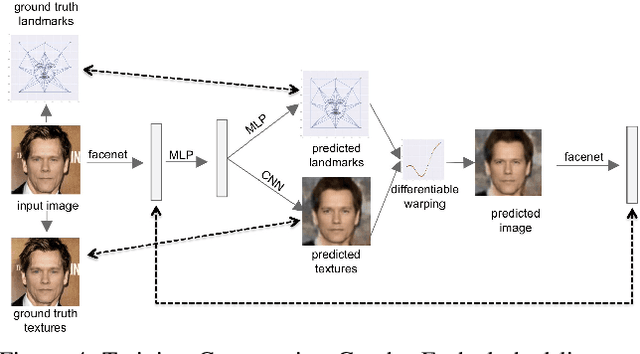
Abstract:We present a method for synthesizing a frontal, neutral-expression image of a person's face given an input face photograph. This is achieved by learning to generate facial landmarks and textures from features extracted from a facial-recognition network. Unlike previous approaches, our encoding feature vector is largely invariant to lighting, pose, and facial expression. Exploiting this invariance, we train our decoder network using only frontal, neutral-expression photographs. Since these photographs are well aligned, we can decompose them into a sparse set of landmark points and aligned texture maps. The decoder then predicts landmarks and textures independently and combines them using a differentiable image warping operation. The resulting images can be used for a number of applications, such as analyzing facial attributes, exposure and white balance adjustment, or creating a 3-D avatar.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge