Electrocardiography Ecg
Papers and Code
ECG-R1: Protocol-Guided and Modality-Agnostic MLLM for Reliable ECG Interpretation
Feb 04, 2026Electrocardiography (ECG) serves as an indispensable diagnostic tool in clinical practice, yet existing multimodal large language models (MLLMs) remain unreliable for ECG interpretation, often producing plausible but clinically incorrect analyses. To address this, we propose ECG-R1, the first reasoning MLLM designed for reliable ECG interpretation via three innovations. First, we construct the interpretation corpus using \textit{Protocol-Guided Instruction Data Generation}, grounding interpretation in measurable ECG features and monograph-defined quantitative thresholds and diagnostic logic. Second, we present a modality-decoupled architecture with \textit{Interleaved Modality Dropout} to improve robustness and cross-modal consistency when either the ECG signal or ECG image is missing. Third, we present \textit{Reinforcement Learning with ECG Diagnostic Evidence Rewards} to strengthen evidence-grounded ECG interpretation. Additionally, we systematically evaluate the ECG interpretation capabilities of proprietary, open-source, and medical MLLMs, and provide the first quantitative evidence that severe hallucinations are widespread, suggesting that the public should not directly trust these outputs without independent verification. Code and data are publicly available at \href{https://github.com/PKUDigitalHealth/ECG-R1}{here}, and an online platform can be accessed at \href{http://ai.heartvoice.com.cn/ECG-R1/}{here}.
Prenatal Stress Detection from Electrocardiography Using Self-Supervised Deep Learning: Development and External Validation
Feb 03, 2026Prenatal psychological stress affects 15-25% of pregnancies and increases risks of preterm birth, low birth weight, and adverse neurodevelopmental outcomes. Current screening relies on subjective questionnaires (PSS-10), limiting continuous monitoring. We developed deep learning models for stress detection from electrocardiography (ECG) using the FELICITy 1 cohort (151 pregnant women, 32-38 weeks gestation). A ResNet-34 encoder was pretrained via SimCLR contrastive learning on 40,692 ECG segments per subject. Multi-layer feature extraction enabled binary classification and continuous PSS prediction across maternal (mECG), fetal (fECG), and abdominal ECG (aECG). External validation used the FELICITy 2 RCT (28 subjects, different ECG device, yoga intervention vs. control). On FELICITy 1 (5-fold CV): mECG 98.6% accuracy (R2=0.88, MAE=1.90), fECG 99.8% (R2=0.95, MAE=1.19), aECG 95.5% (R2=0.75, MAE=2.80). External validation on FELICITy 2: mECG 77.3% accuracy (R2=0.62, MAE=3.54, AUC=0.826), aECG 63.6% (R2=0.29, AUC=0.705). Signal quality-based channel selection outperformed all-channel averaging (+12% R2 improvement). Mixed-effects models detected a significant intervention response (p=0.041). Self-supervised deep learning on pregnancy ECG enables accurate, objective stress assessment, with multi-layer feature extraction substantially outperforming single embedding approaches.
AnyECG: Evolved ECG Foundation Model for Holistic Health Profiling
Jan 12, 2026Background: Artificial intelligence enabled electrocardiography (AI-ECG) has demonstrated the ability to detect diverse pathologies, but most existing models focus on single disease identification, neglecting comorbidities and future risk prediction. Although ECGFounder expanded cardiac disease coverage, a holistic health profiling model remains needed. Methods: We constructed a large multicenter dataset comprising 13.3 million ECGs from 2.98 million patients. Using transfer learning, ECGFounder was fine-tuned to develop AnyECG, a foundation model for holistic health profiling. Performance was evaluated using external validation cohorts and a 10-year longitudinal cohort for current diagnosis, future risk prediction, and comorbidity identification. Results: AnyECG demonstrated systemic predictive capability across 1172 conditions, achieving an AUROC greater than 0.7 for 306 diseases. The model revealed novel disease associations, robust comorbidity patterns, and future disease risks. Representative examples included high diagnostic performance for hyperparathyroidism (AUROC 0.941), type 2 diabetes (0.803), Crohn disease (0.817), lymphoid leukemia (0.856), and chronic obstructive pulmonary disease (0.773). Conclusion: The AnyECG foundation model provides substantial evidence that AI-ECG can serve as a systemic tool for concurrent disease detection and long-term risk prediction.
Hear the Heartbeat in Phases: Physiologically Grounded Phase-Aware ECG Biometrics
Jan 01, 2026Electrocardiography (ECG) is adopted for identity authentication in wearable devices due to its individual-specific characteristics and inherent liveness. However, existing methods often treat heartbeats as homogeneous signals, overlooking the phase-specific characteristics within the cardiac cycle. To address this, we propose a Hierarchical Phase-Aware Fusion~(HPAF) framework that explicitly avoids cross-feature entanglement through a three-stage design. In the first stage, Intra-Phase Representation (IPR) independently extracts representations for each cardiac phase, ensuring that phase-specific morphological and variation cues are preserved without interference from other phases. In the second stage, Phase-Grouped Hierarchical Fusion (PGHF) aggregates physiologically related phases in a structured manner, enabling reliable integration of complementary phase information. In the final stage, Global Representation Fusion (GRF) further combines the grouped representations and adaptively balances their contributions to produce a unified and discriminative identity representation. Moreover, considering ECG signals are continuously acquired, multiple heartbeats can be collected for each individual. We propose a Heartbeat-Aware Multi-prototype (HAM) enrollment strategy, which constructs a multi-prototype gallery template set to reduce the impact of heartbeat-specific noise and variability. Extensive experiments on three public datasets demonstrate that HPAF achieves state-of-the-art results in the comparison with other methods under both closed and open-set settings.
Human-like visual computing advances explainability and few-shot learning in deep neural networks for complex physiological data
Dec 26, 2025Machine vision models, particularly deep neural networks, are increasingly applied to physiological signal interpretation, including electrocardiography (ECG), yet they typically require large training datasets and offer limited insight into the causal features underlying their predictions. This lack of data efficiency and interpretability constrains their clinical reliability and alignment with human reasoning. Here, we show that a perception-informed pseudo-colouring technique, previously demonstrated to enhance human ECG interpretation, can improve both explainability and few-shot learning in deep neural networks analysing complex physiological data. We focus on acquired, drug-induced long QT syndrome (LQTS) as a challenging case study characterised by heterogeneous signal morphology, variable heart rate, and scarce positive cases associated with life-threatening arrhythmias such as torsades de pointes. This setting provides a stringent test of model generalisation under extreme data scarcity. By encoding clinically salient temporal features, such as QT-interval duration, into structured colour representations, models learn discriminative and interpretable features from as few as one or five training examples. Using prototypical networks and a ResNet-18 architecture, we evaluate one-shot and few-shot learning on ECG images derived from single cardiac cycles and full 10-second rhythms. Explainability analyses show that pseudo-colouring guides attention toward clinically meaningful ECG features while suppressing irrelevant signal components. Aggregating multiple cardiac cycles further improves performance, mirroring human perceptual averaging across heartbeats. Together, these findings demonstrate that human-like perceptual encoding can bridge data efficiency, explainability, and causal reasoning in medical machine intelligence.
AnyECG-Lab: An Exploration Study of Fine-tuning an ECG Foundation Model to Estimate Laboratory Values from Single-Lead ECG Signals
Oct 25, 2025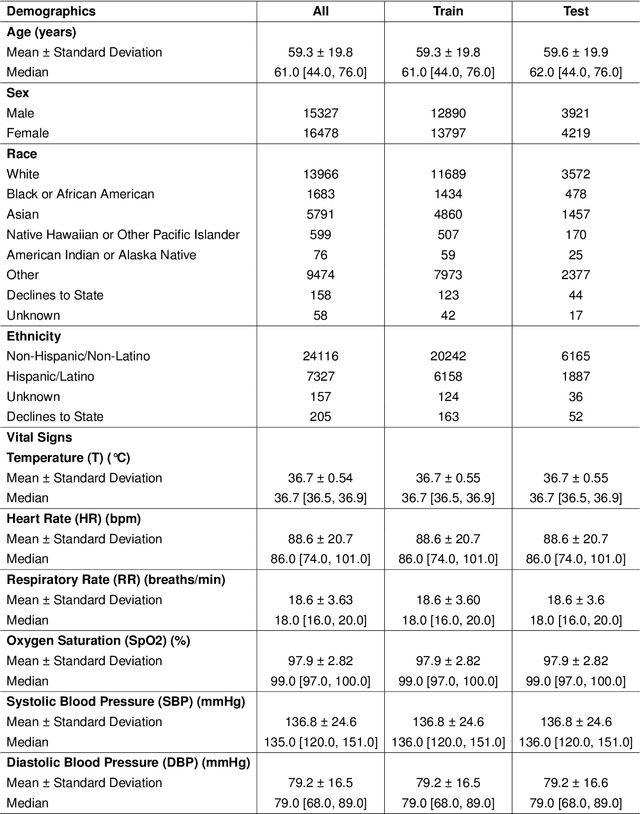
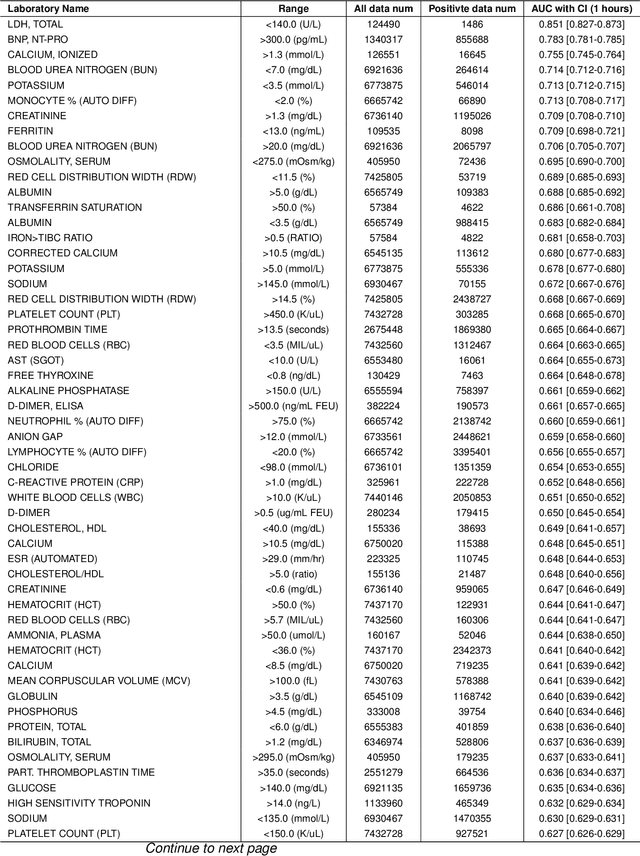
Timely access to laboratory values is critical for clinical decision-making, yet current approaches rely on invasive venous sampling and are intrinsically delayed. Electrocardiography (ECG), as a non-invasive and widely available signal, offers a promising modality for rapid laboratory estimation. Recent progress in deep learning has enabled the extraction of latent hematological signatures from ECGs. However, existing models are constrained by low signal-to-noise ratios, substantial inter-individual variability, limited data diversity, and suboptimal generalization, especially when adapted to low-lead wearable devices. In this work, we conduct an exploratory study leveraging transfer learning to fine-tune ECGFounder, a large-scale pre-trained ECG foundation model, on the Multimodal Clinical Monitoring in the Emergency Department (MC-MED) dataset from Stanford. We generated a corpus of more than 20 million standardized ten-second ECG segments to enhance sensitivity to subtle biochemical correlates. On internal validation, the model demonstrated strong predictive performance (area under the curve above 0.65) for thirty-three laboratory indicators, moderate performance (between 0.55 and 0.65) for fifty-nine indicators, and limited performance (below 0.55) for sixteen indicators. This study provides an efficient artificial-intelligence driven solution and establishes the feasibility scope for real-time, non-invasive estimation of laboratory values.
MultiPhysio-HRC: Multimodal Physiological Signals Dataset for industrial Human-Robot Collaboration
Oct 01, 2025Human-robot collaboration (HRC) is a key focus of Industry 5.0, aiming to enhance worker productivity while ensuring well-being. The ability to perceive human psycho-physical states, such as stress and cognitive load, is crucial for adaptive and human-aware robotics. This paper introduces MultiPhysio-HRC, a multimodal dataset containing physiological, audio, and facial data collected during real-world HRC scenarios. The dataset includes electroencephalography (EEG), electrocardiography (ECG), electrodermal activity (EDA), respiration (RESP), electromyography (EMG), voice recordings, and facial action units. The dataset integrates controlled cognitive tasks, immersive virtual reality experiences, and industrial disassembly activities performed manually and with robotic assistance, to capture a holistic view of the participants' mental states. Rich ground truth annotations were obtained using validated psychological self-assessment questionnaires. Baseline models were evaluated for stress and cognitive load classification, demonstrating the dataset's potential for affective computing and human-aware robotics research. MultiPhysio-HRC is publicly available to support research in human-centered automation, workplace well-being, and intelligent robotic systems.
Domain Knowledge is Power: Leveraging Physiological Priors for Self Supervised Representation Learning in Electrocardiography
Sep 09, 2025Objective: Electrocardiograms (ECGs) play a crucial role in diagnosing heart conditions; however, the effectiveness of artificial intelligence (AI)-based ECG analysis is often hindered by the limited availability of labeled data. Self-supervised learning (SSL) can address this by leveraging large-scale unlabeled data. We introduce PhysioCLR (Physiology-aware Contrastive Learning Representation for ECG), a physiology-aware contrastive learning framework that incorporates domain-specific priors to enhance the generalizability and clinical relevance of ECG-based arrhythmia classification. Methods: During pretraining, PhysioCLR learns to bring together embeddings of samples that share similar clinically relevant features while pushing apart those that are dissimilar. Unlike existing methods, our method integrates ECG physiological similarity cues into contrastive learning, promoting the learning of clinically meaningful representations. Additionally, we introduce ECG- specific augmentations that preserve the ECG category post augmentation and propose a hybrid loss function to further refine the quality of learned representations. Results: We evaluate PhysioCLR on two public ECG datasets, Chapman and Georgia, for multilabel ECG diagnoses, as well as a private ICU dataset labeled for binary classification. Across the Chapman, Georgia, and private cohorts, PhysioCLR boosts the mean AUROC by 12% relative to the strongest baseline, underscoring its robust cross-dataset generalization. Conclusion: By embedding physiological knowledge into contrastive learning, PhysioCLR enables the model to learn clinically meaningful and transferable ECG eatures. Significance: PhysioCLR demonstrates the potential of physiology-informed SSL to offer a promising path toward more effective and label-efficient ECG diagnostics.
Comprehensive Signal Quality Evaluation of a Wearable Textile ECG Garment: A Sex-Balanced Study
Aug 29, 2025We introduce a novel wearable textile-garment featuring an innovative electrode placement aimed at minimizing noise and motion artifacts, thereby enhancing signal fidelity in Electrocardiography (ECG) recordings. We present a comprehensive, sex-balanced evaluation involving 15 healthy males and 15 healthy female participants to ensure the device's suitability across anatomical and physiological variations. The assessment framework encompasses distinct evaluation approaches: quantitative signal quality indices to objectively benchmark device performance; rhythm-based analyzes of physiological parameters such as heart rate and heart rate variability; machine learning classification tasks to assess application-relevant predictive utility; morphological analysis of ECG features including amplitude and interval parameters; and investigations of the effects of electrode projection angle given by the textile / body shape, with all analyzes stratified by sex to elucidate sex-specific influences. Evaluations were conducted across various activity phases representing real-world conditions. The results demonstrate that the textile system achieves signal quality highly concordant with reference devices in both rhythm and morphological analyses, exhibits robust classification performance, and enables identification of key sex-specific determinants affecting signal acquisition. These findings underscore the practical viability of textile-based ECG garments for physiological monitoring as well as psychophysiological state detection. Moreover, we identify the importance of incorporating sex-specific design considerations to ensure equitable and reliable cardiac diagnostics in wearable health technologies.
Diffusion-Based Electrocardiography Noise Quantification via Anomaly Detection
Jun 13, 2025Electrocardiography (ECG) signals are often degraded by noise, which complicates diagnosis in clinical and wearable settings. This study proposes a diffusion-based framework for ECG noise quantification via reconstruction-based anomaly detection, addressing annotation inconsistencies and the limited generalizability of conventional methods. We introduce a distributional evaluation using the Wasserstein-1 distance ($W_1$), comparing the reconstruction error distributions between clean and noisy ECGs to mitigate inconsistent annotations. Our final model achieved robust noise quantification using only three reverse diffusion steps. The model recorded a macro-average $W_1$ score of 1.308 across the benchmarks, outperforming the next-best method by over 48%. External validations demonstrated strong generalizability, supporting the exclusion of low-quality segments to enhance diagnostic accuracy and enable timely clinical responses to signal degradation. The proposed method enhances clinical decision-making, diagnostic accuracy, and real-time ECG monitoring capabilities, supporting future advancements in clinical and wearable ECG applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge