Yutao Ma
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025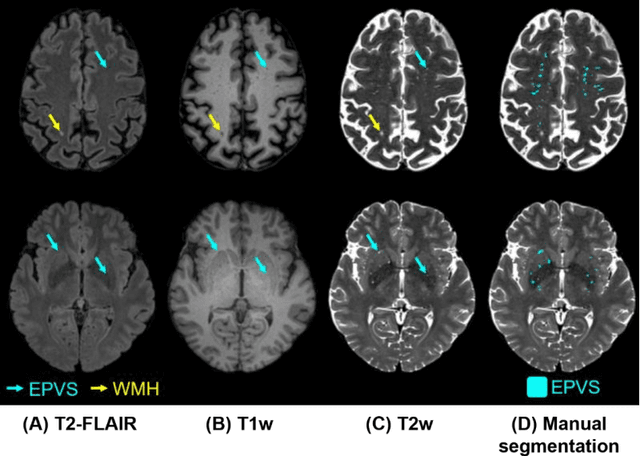
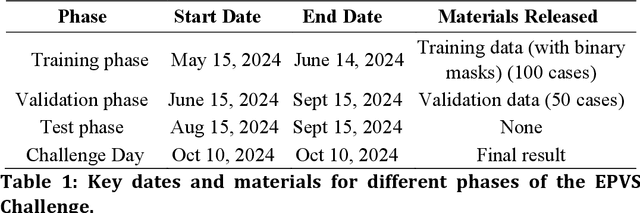
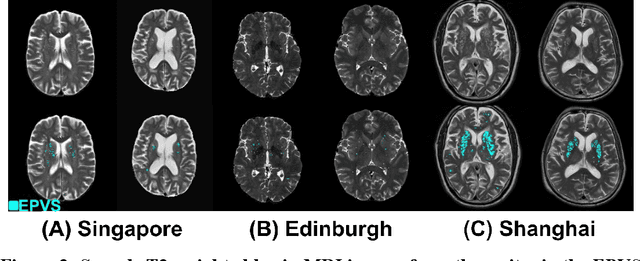
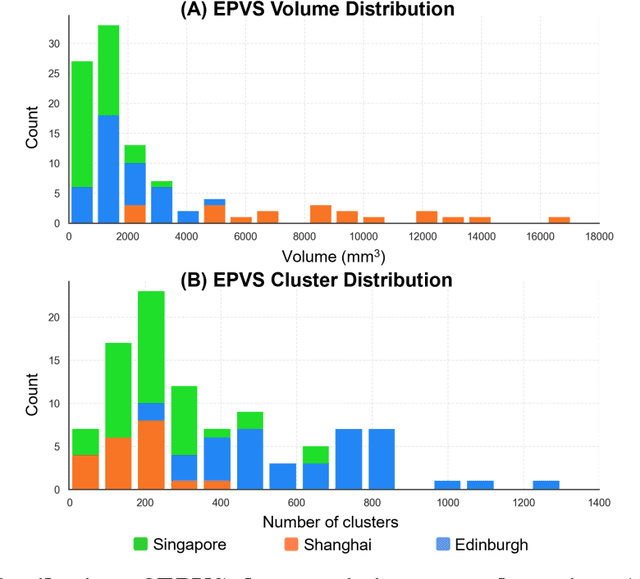
Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Fine-Tuning Code Language Models to Detect Cross-Language Bugs
Jul 29, 2025Abstract:Multilingual programming, which involves using multiple programming languages (PLs) in a single project, is increasingly common due to its benefits. However, it introduces cross-language bugs (CLBs), which arise from interactions between different PLs and are difficult to detect by single-language bug detection tools. This paper investigates the potential of pre-trained code language models (CodeLMs) in CLB detection. We developed CLCFinder, a cross-language code identification tool, and constructed a CLB dataset involving three PL combinations (Python-C/C++, Java-C/C++, and Python-Java) with nine interaction types. We fine-tuned 13 CodeLMs on this dataset and evaluated their performance, analyzing the effects of dataset size, token sequence length, and code comments. Results show that all CodeLMs performed poorly before fine-tuning, but exhibited varying degrees of performance improvement after fine-tuning, with UniXcoder-base achieving the best F1 score (0.7407). Notably, small fine-tuned CodeLMs tended to performe better than large ones. CodeLMs fine-tuned on single-language bug datasets performed poorly on CLB detection, demonstrating the distinction between CLBs and single-language bugs. Additionally, increasing the fine-tuning dataset size significantly improved performance, while longer token sequences did not necessarily improve the model performance. The impact of code comments varied across models. Some fine-tuned CodeLMs' performance was improved, while others showed degraded performance.
SciCode: A Research Coding Benchmark Curated by Scientists
Jul 18, 2024



Abstract:Since language models (LMs) now outperform average humans on many challenging tasks, it has become increasingly difficult to develop challenging, high-quality, and realistic evaluations. We address this issue by examining LMs' capabilities to generate code for solving real scientific research problems. Incorporating input from scientists and AI researchers in 16 diverse natural science sub-fields, including mathematics, physics, chemistry, biology, and materials science, we created a scientist-curated coding benchmark, SciCode. The problems in SciCode naturally factorize into multiple subproblems, each involving knowledge recall, reasoning, and code synthesis. In total, SciCode contains 338 subproblems decomposed from 80 challenging main problems. It offers optional descriptions specifying useful scientific background information and scientist-annotated gold-standard solutions and test cases for evaluation. Claude3.5-Sonnet, the best-performing model among those tested, can solve only 4.6% of the problems in the most realistic setting. We believe that SciCode demonstrates both contemporary LMs' progress towards becoming helpful scientific assistants and sheds light on the development and evaluation of scientific AI in the future.
Cervical Optical Coherence Tomography Image Classification Based on Contrastive Self-Supervised Texture Learning
Aug 11, 2021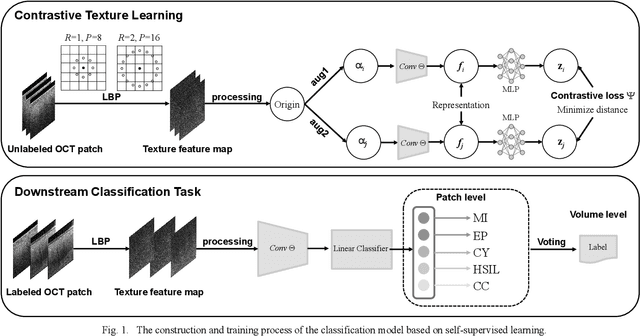
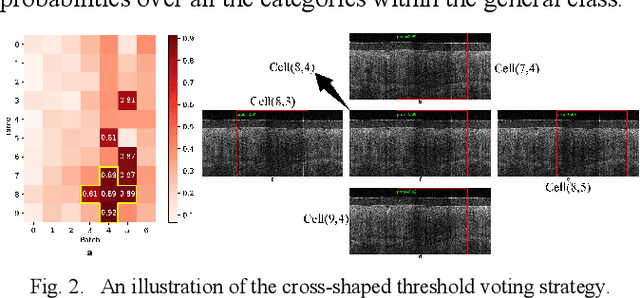
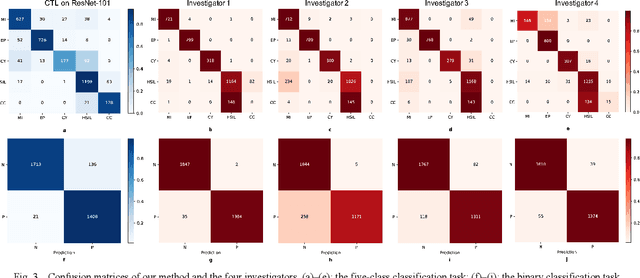
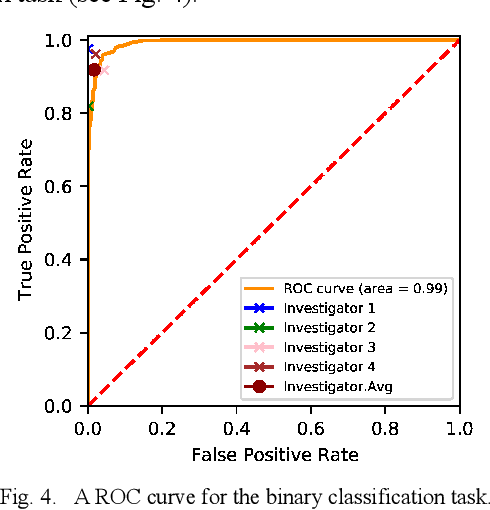
Abstract:Background: Cervical cancer seriously affects the health of the female reproductive system. Optical coherence tomography (OCT) emerges as a non-invasive, high-resolution imaging technology for cervical disease detection. However, OCT image annotation is knowledge-intensive and time-consuming, which impedes the training process of deep-learning-based classification models. Objective: This study aims to develop a computer-aided diagnosis (CADx) approach to classifying in-vivo cervical OCT images based on self-supervised learning. Methods: Besides high-level semantic features extracted by a convolutional neural network (CNN), the proposed CADx approach leverages unlabeled cervical OCT images' texture features learned by contrastive texture learning. We conducted ten-fold cross-validation on the OCT image dataset from a multi-center clinical study on 733 patients from China. Results: In a binary classification task for detecting high-risk diseases, including high-grade squamous intraepithelial lesion (HSIL) and cervical cancer, our method achieved an area-under-the-curve (AUC) value of 0.9798 Plus or Minus 0.0157 with a sensitivity of 91.17 Plus or Minus 4.99% and a specificity of 93.96 Plus or Minus 4.72% for OCT image patches; also, it outperformed two out of four medical experts on the test set. Furthermore, our method achieved a 91.53% sensitivity and 97.37% specificity on an external validation dataset containing 287 3D OCT volumes from 118 Chinese patients in a new hospital using a cross-shaped threshold voting strategy. Conclusion: The proposed contrastive-learning-based CADx method outperformed the end-to-end CNN models and provided better interpretability based on texture features, which holds great potential to be used in the clinical protocol of "see-and-treat."
Position-enhanced and Time-aware Graph Convolutional Network for Sequential Recommendations
Jul 12, 2021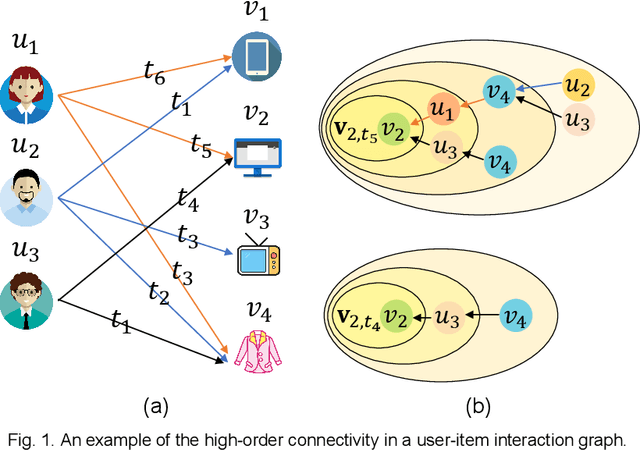
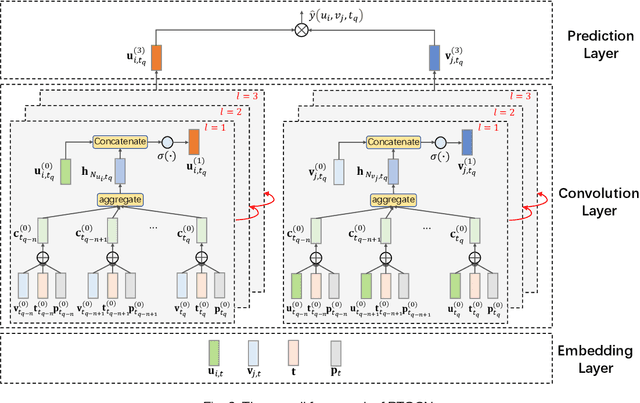
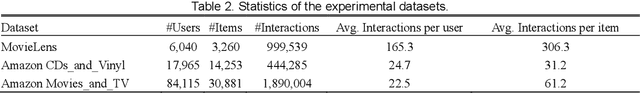
Abstract:Most of the existing deep learning-based sequential recommendation approaches utilize the recurrent neural network architecture or self-attention to model the sequential patterns and temporal influence among a user's historical behavior and learn the user's preference at a specific time. However, these methods have two main drawbacks. First, they focus on modeling users' dynamic states from a user-centric perspective and always neglect the dynamics of items over time. Second, most of them deal with only the first-order user-item interactions and do not consider the high-order connectivity between users and items, which has recently been proved helpful for the sequential recommendation. To address the above problems, in this article, we attempt to model user-item interactions by a bipartite graph structure and propose a new recommendation approach based on a Position-enhanced and Time-aware Graph Convolutional Network (PTGCN) for the sequential recommendation. PTGCN models the sequential patterns and temporal dynamics between user-item interactions by defining a position-enhanced and time-aware graph convolution operation and learning the dynamic representations of users and items simultaneously on the bipartite graph with a self-attention aggregator. Also, it realizes the high-order connectivity between users and items by stacking multi-layer graph convolutions. To demonstrate the effectiveness of PTGCN, we carried out a comprehensive evaluation of PTGCN on three real-world datasets of different sizes compared with a few competitive baselines. Experimental results indicate that PTGCN outperforms several state-of-the-art models in terms of two commonly-used evaluation metrics for ranking.
Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms
Apr 24, 2019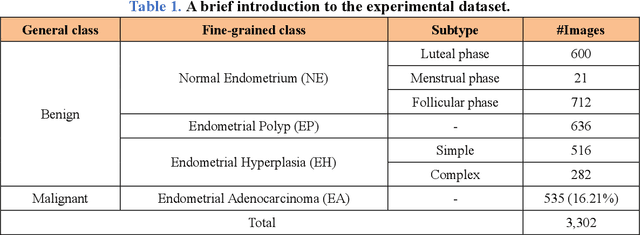
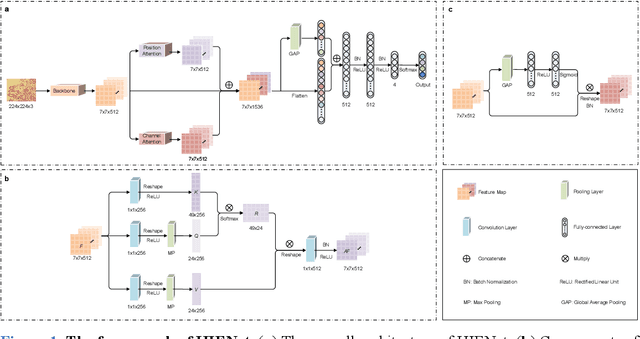
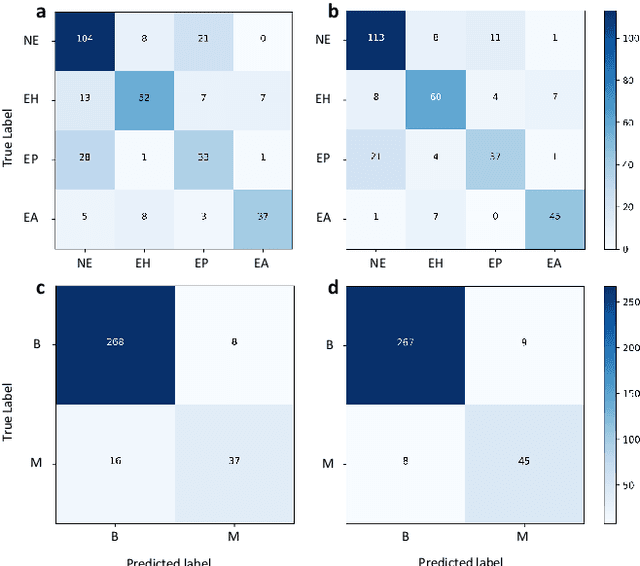
Abstract:Uterine cancer, also known as endometrial cancer, can seriously affect the female reproductive organs, and histopathological image analysis is the gold standard for diagnosing endometrial cancer. However, due to the limited capability of modeling the complicated relationships between histopathological images and their interpretations, these computer-aided diagnosis (CADx) approaches based on traditional machine learning algorithms often failed to achieve satisfying results. In this study, we developed a CADx approach using a convolutional neural network (CNN) and attention mechanisms, called HIENet. Because HIENet used the attention mechanisms and feature map visualization techniques, it can provide pathologists better interpretability of diagnoses by highlighting the histopathological correlations of local (pixel-level) image features to morphological characteristics of endometrial tissue. In the ten-fold cross-validation process, the CADx approach, HIENet, achieved a 76.91 $\pm$ 1.17% (mean $\pm$ s. d.) classification accuracy for four classes of endometrial tissue, namely normal endometrium, endometrial polyp, endometrial hyperplasia, and endometrial adenocarcinoma. Also, HIENet achieved an area-under-the-curve (AUC) of 0.9579 $\pm$ 0.0103 with an 81.04 $\pm$ 3.87% sensitivity and 94.78 $\pm$ 0.87% specificity in a binary classification task that detected endometrioid adenocarcinoma (Malignant). Besides, in the external validation process, HIENet achieved an 84.50% accuracy in the four-class classification task, and it achieved an AUC of 0.9829 with a 77.97% (95% CI, 65.27%-87.71%) sensitivity and 100% (95% CI, 97.42%-100.00%) specificity. In summary, the proposed CADx approach, HIENet, outperformed three human experts and four end-to-end CNN-based classifiers on this small-scale dataset composed of 3,500 hematoxylin and eosin (H&E) images regarding overall classification performance.
Computer-Aided Diagnosis of Label-Free 3-D Optical Coherence Microscopy Images of Human Cervical Tissue
Sep 17, 2018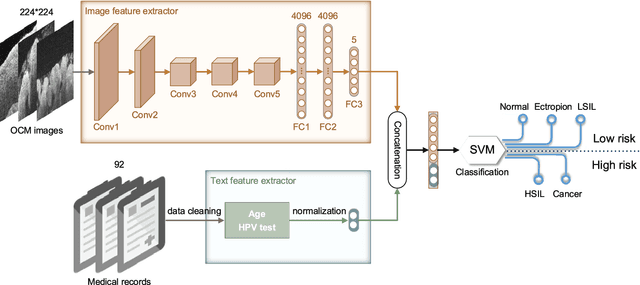
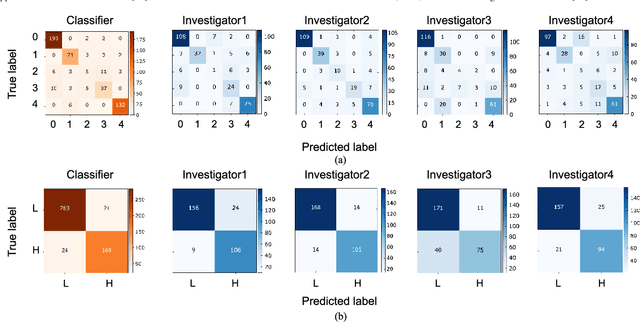
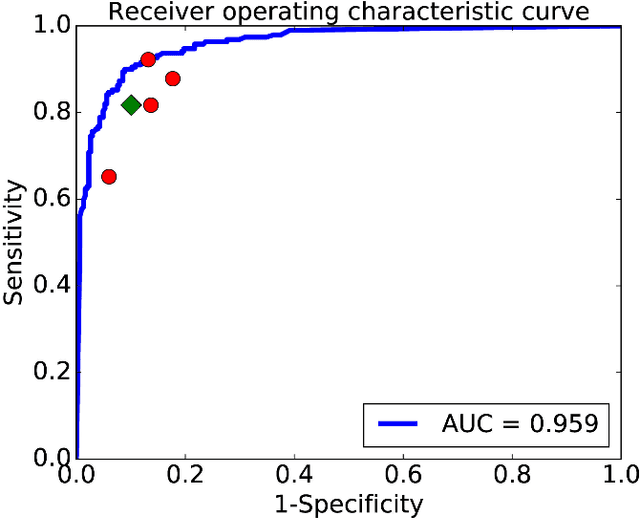
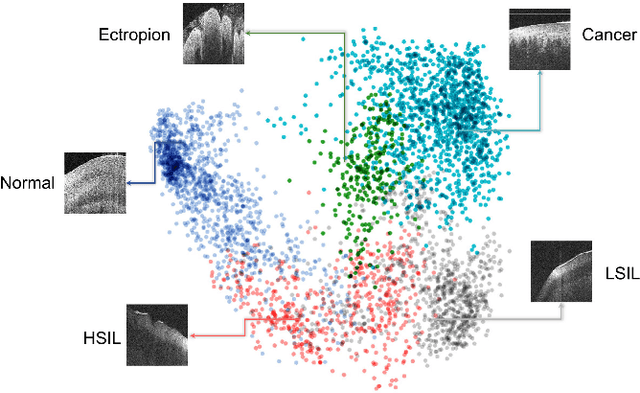
Abstract:Objective: Ultrahigh-resolution optical coherence microscopy (OCM) has recently demonstrated its potential for accurate diagnosis of human cervical diseases. One major challenge for clinical adoption, however, is the steep learning curve clinicians need to overcome to interpret OCM images. Developing an intelligent technique for computer-aided diagnosis (CADx) to accurately interpret OCM images will facilitate clinical adoption of the technology and improve patient care. Methods: 497 high-resolution 3-D OCM volumes (600 cross-sectional images each) were collected from 159 ex vivo specimens of 92 female patients. OCM image features were extracted using a convolutional neural network (CNN) model, concatenated with patient information (e.g., age, HPV results), and classified using a support vector machine classifier. Ten-fold cross-validations were utilized to test the performance of the CADx method in a five-class classification task and a binary classification task. Results: An 88.3 plus or minus 4.9% classification accuracy was achieved for five fine-grained classes of cervical tissue, namely normal, ectropion, low-grade and high-grade squamous intraepithelial lesions (LSIL and HSIL), and cancer. In the binary classification task (low-risk [normal, ectropion and LSIL] vs. high-risk [HSIL and cancer]), the CADx method achieved an area-under-the-curve (AUC) value of 0.959 with an 86.7 plus or minus 11.4% sensitivity and 93.5 plus or minus 3.8% specificity. Conclusion: The proposed deep-learning based CADx method outperformed three human experts. It was also able to identify morphological characteristics in OCM images that were consistent with histopathological interpretations. Significance: Label-free OCM imaging, combined with deep-learning based CADx methods, hold a great promise to be used in clinical settings for the effective screening and diagnosis of cervical diseases.
Segmentation of Drosophila Heart in Optical Coherence Microscopy Images Using Convolutional Neural Networks
Jul 21, 2018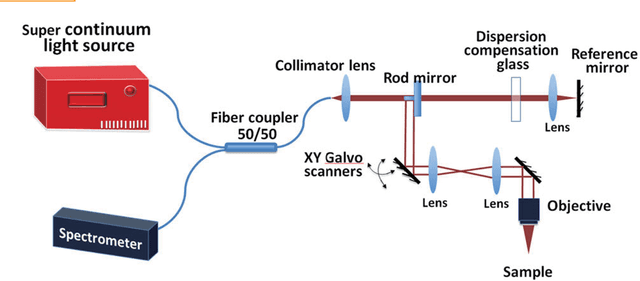

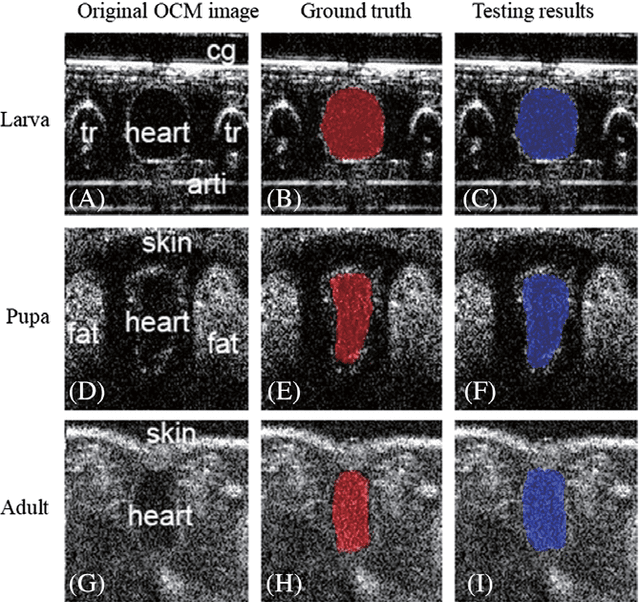
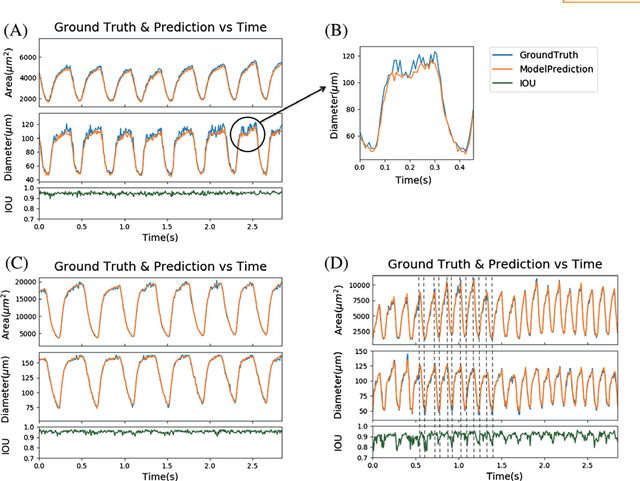
Abstract:Convolutional neural networks are powerful tools for image segmentation and classification. Here, we use this method to identify and mark the heart region of Drosophila at different developmental stages in the cross-sectional images acquired by a custom optical coherence microscopy (OCM) system. With our well-trained convolutional neural network model, the heart regions through multiple heartbeat cycles can be marked with an intersection over union (IOU) of ~86%. Various morphological and dynamical cardiac parameters can be quantified accurately with automatically segmented heart regions. This study demonstrates an efficient heart segmentation method to analyze OCM images of the beating heart in Drosophila.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge