Yulia Otmakhova
LLMs for Argument Mining: Detection, Extraction, and Relationship Classification of pre-defined Arguments in Online Comments
May 29, 2025



Abstract:Automated large-scale analysis of public discussions around contested issues like abortion requires detecting and understanding the use of arguments. While Large Language Models (LLMs) have shown promise in language processing tasks, their performance in mining topic-specific, pre-defined arguments in online comments remains underexplored. We evaluate four state-of-the-art LLMs on three argument mining tasks using datasets comprising over 2,000 opinion comments across six polarizing topics. Quantitative evaluation suggests an overall strong performance across the three tasks, especially for large and fine-tuned LLMs, albeit at a significant environmental cost. However, a detailed error analysis revealed systematic shortcomings on long and nuanced comments and emotionally charged language, raising concerns for downstream applications like content moderation or opinion analysis. Our results highlight both the promise and current limitations of LLMs for automated argument analysis in online comments.
FLUKE: A Linguistically-Driven and Task-Agnostic Framework for Robustness Evaluation
Apr 24, 2025



Abstract:We present FLUKE (Framework for LingUistically-driven and tasK-agnostic robustness Evaluation), a task-agnostic framework for assessing model robustness through systematic minimal variations of test data. FLUKE introduces controlled variations across linguistic levels - from orthography to dialect and style varieties - and leverages large language models (LLMs) with human validation to generate modifications. We demonstrate FLUKE's utility by evaluating both fine-tuned models and LLMs across four diverse NLP tasks, and reveal that (1) the impact of linguistic variations is highly task-dependent, with some tests being critical for certain tasks but irrelevant for others; (2) while LLMs have better overall robustness compared to fine-tuned models, they still exhibit significant brittleness to certain linguistic variations; (3) all models show substantial vulnerability to negation modifications across most tasks. These findings highlight the importance of systematic robustness testing for understanding model behaviors.
Generative Debunking of Climate Misinformation
Jul 08, 2024Abstract:Misinformation about climate change causes numerous negative impacts, necessitating corrective responses. Psychological research has offered various strategies for reducing the influence of climate misinformation, such as the fact-myth-fallacy-fact-structure. However, practically implementing corrective interventions at scale represents a challenge. Automatic detection and correction of misinformation offers a solution to the misinformation problem. This study documents the development of large language models that accept as input a climate myth and produce a debunking that adheres to the fact-myth-fallacy-fact (``truth sandwich'') structure, by incorporating contrarian claim classification and fallacy detection into an LLM prompting framework. We combine open (Mixtral, Palm2) and proprietary (GPT-4) LLMs with prompting strategies of varying complexity. Experiments reveal promising performance of GPT-4 and Mixtral if combined with structured prompts. We identify specific challenges of debunking generation and human evaluation, and map out avenues for future work. We release a dataset of high-quality truth-sandwich debunkings, source code and a demo of the debunking system.
Revisiting subword tokenization: A case study on affixal negation in large language models
Apr 04, 2024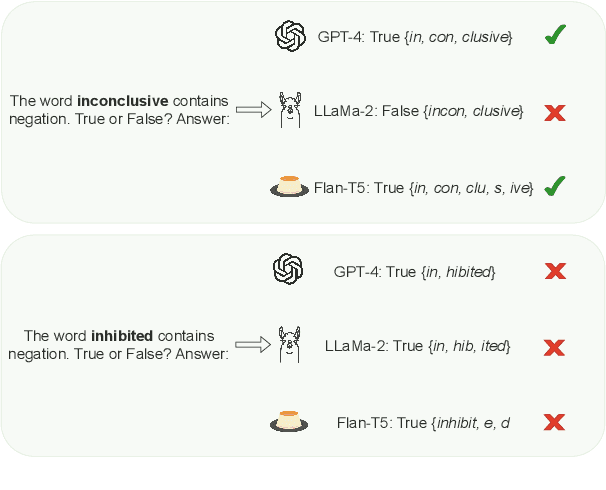
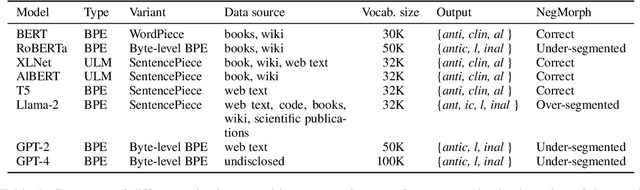
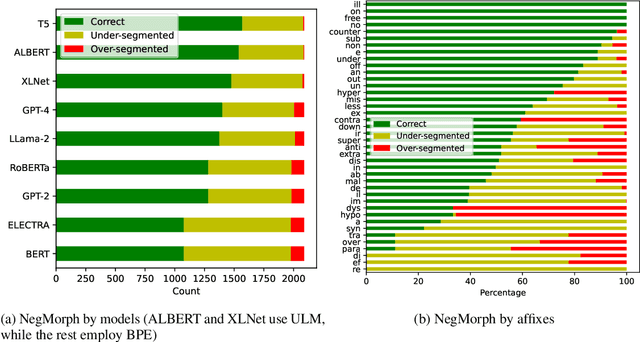
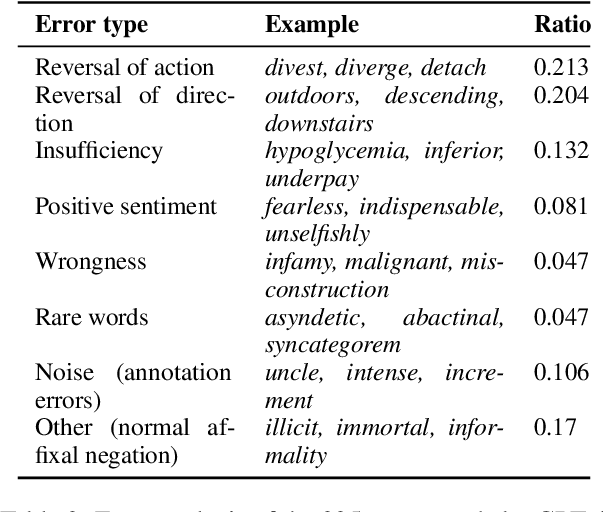
Abstract:In this work, we measure the impact of affixal negation on modern English large language models (LLMs). In affixal negation, the negated meaning is expressed through a negative morpheme, which is potentially challenging for LLMs as their tokenizers are often not morphologically plausible. We conduct extensive experiments using LLMs with different subword tokenization methods, which lead to several insights on the interaction between tokenization performance and negation sensitivity. Despite some interesting mismatches between tokenization accuracy and negation detection performance, we show that models can, on the whole, reliably recognize the meaning of affixal negation.
Automated Metrics for Medical Multi-Document Summarization Disagree with Human Evaluations
May 23, 2023Abstract:Evaluating multi-document summarization (MDS) quality is difficult. This is especially true in the case of MDS for biomedical literature reviews, where models must synthesize contradicting evidence reported across different documents. Prior work has shown that rather than performing the task, models may exploit shortcuts that are difficult to detect using standard n-gram similarity metrics such as ROUGE. Better automated evaluation metrics are needed, but few resources exist to assess metrics when they are proposed. Therefore, we introduce a dataset of human-assessed summary quality facets and pairwise preferences to encourage and support the development of better automated evaluation methods for literature review MDS. We take advantage of community submissions to the Multi-document Summarization for Literature Review (MSLR) shared task to compile a diverse and representative sample of generated summaries. We analyze how automated summarization evaluation metrics correlate with lexical features of generated summaries, to other automated metrics including several we propose in this work, and to aspects of human-assessed summary quality. We find that not only do automated metrics fail to capture aspects of quality as assessed by humans, in many cases the system rankings produced by these metrics are anti-correlated with rankings according to human annotators.
Not another Negation Benchmark: The NaN-NLI Test Suite for Sub-clausal Negation
Oct 06, 2022
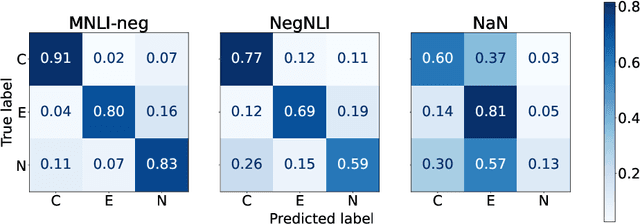
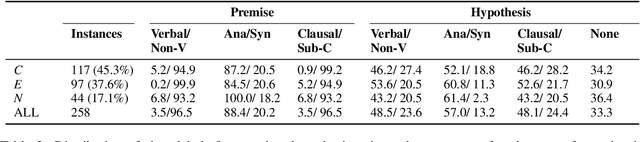
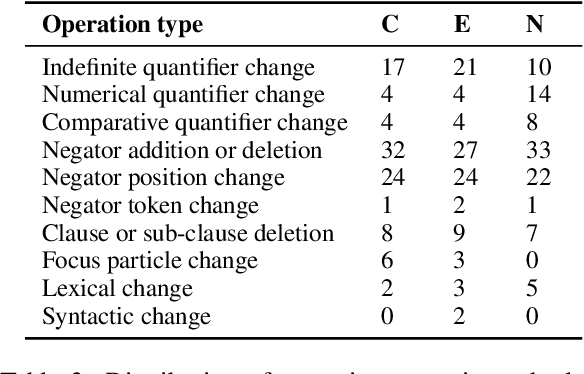
Abstract:Negation is poorly captured by current language models, although the extent of this problem is not widely understood. We introduce a natural language inference (NLI) test suite to enable probing the capabilities of NLP methods, with the aim of understanding sub-clausal negation. The test suite contains premise--hypothesis pairs where the premise contains sub-clausal negation and the hypothesis is constructed by making minimal modifications to the premise in order to reflect different possible interpretations. Aside from adopting standard NLI labels, our test suite is systematically constructed under a rigorous linguistic framework. It includes annotation of negation types and constructions grounded in linguistic theory, as well as the operations used to construct hypotheses. This facilitates fine-grained analysis of model performance. We conduct experiments using pre-trained language models to demonstrate that our test suite is more challenging than existing benchmarks focused on negation, and show how our annotation supports a deeper understanding of the current NLI capabilities in terms of negation and quantification.
LED down the rabbit hole: exploring the potential of global attention for biomedical multi-document summarisation
Sep 19, 2022
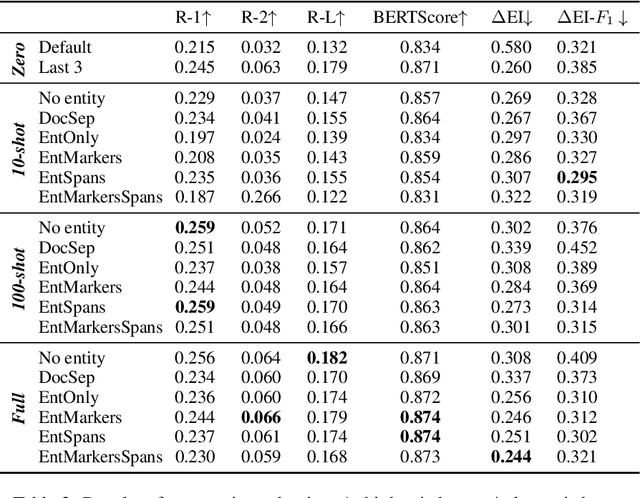
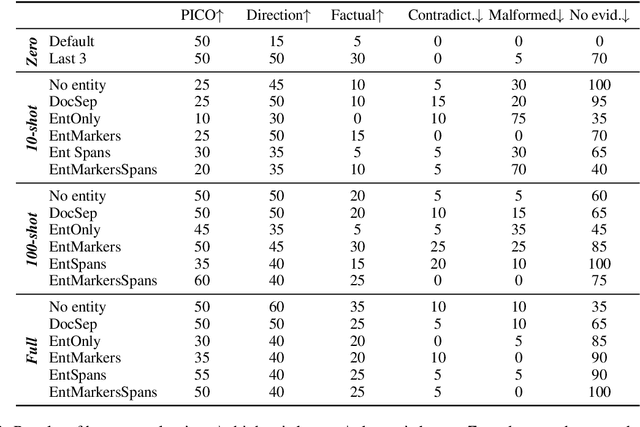
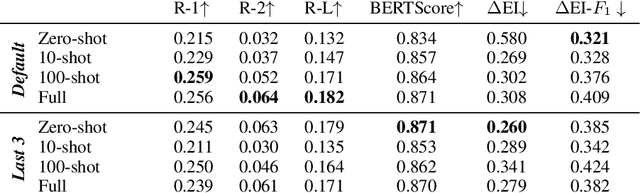
Abstract:In this paper we report on our submission to the Multidocument Summarisation for Literature Review (MSLR) shared task. Specifically, we adapt PRIMERA (Xiao et al., 2022) to the biomedical domain by placing global attention on important biomedical entities in several ways. We analyse the outputs of the 23 resulting models, and report patterns in the results related to the presence of additional global attention, number of training steps, and the input configuration.
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022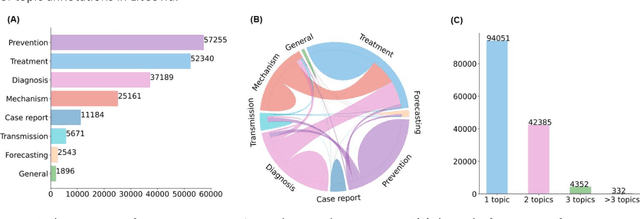
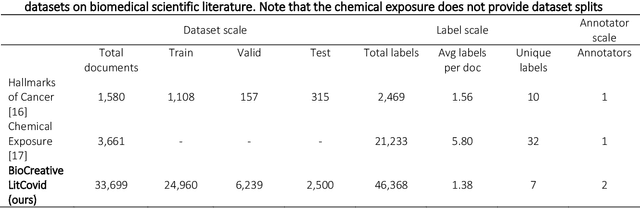
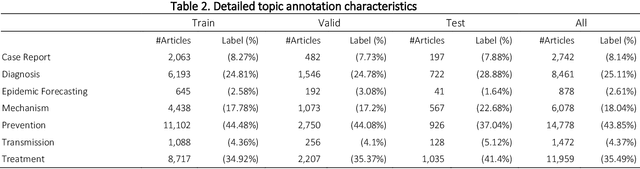
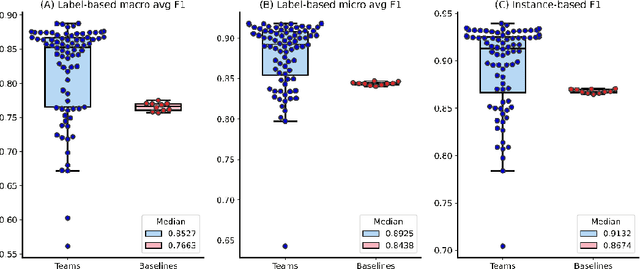
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
ITTC @ TREC 2021 Clinical Trials Track
Feb 16, 2022
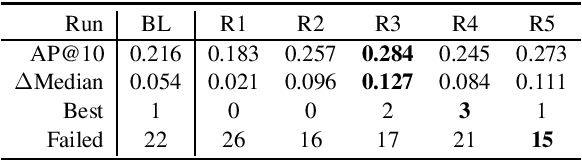
Abstract:This paper describes the submissions of the Natural Language Processing (NLP) team from the Australian Research Council Industrial Transformation Training Centre (ITTC) for Cognitive Computing in Medical Technologies to the TREC 2021 Clinical Trials Track. The task focuses on the problem of matching eligible clinical trials to topics constituting a summary of a patient's admission notes. We explore different ways of representing trials and topics using NLP techniques, and then use a common retrieval model to generate the ranked list of relevant trials for each topic. The results from all our submitted runs are well above the median scores for all topics, but there is still plenty of scope for improvement.
Impact of detecting clinical trial elements in exploration of COVID-19 literature
May 25, 2021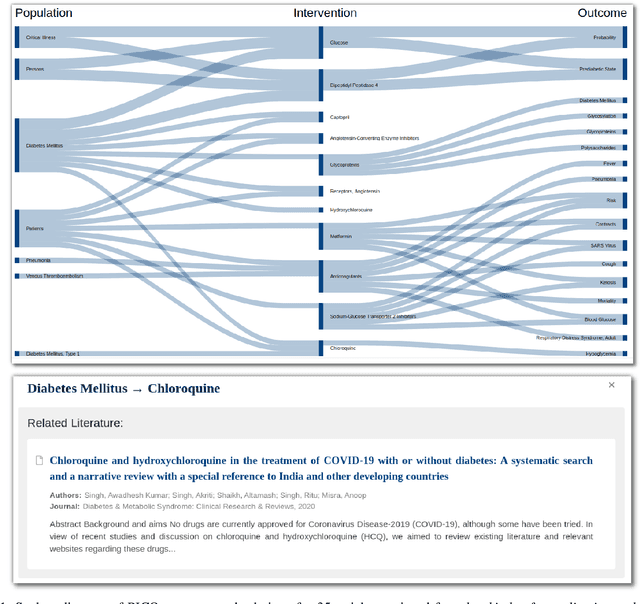
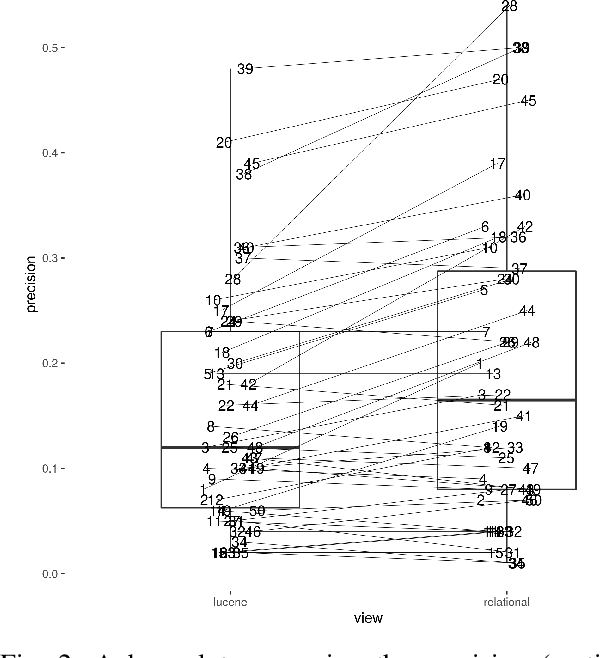
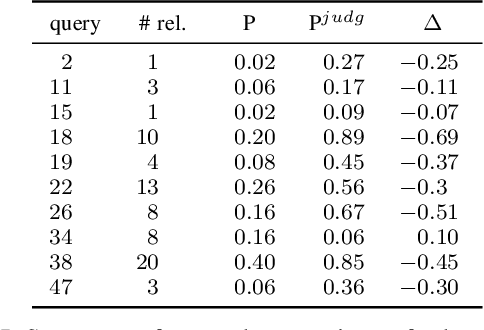

Abstract:The COVID-19 pandemic has driven ever-greater demand for tools which enable efficient exploration of biomedical literature. Although semi-structured information resulting from concept recognition and detection of the defining elements of clinical trials (e.g. PICO criteria) has been commonly used to support literature search, the contributions of this abstraction remain poorly understood, especially in relation to text-based retrieval. In this study, we compare the results retrieved by a standard search engine with those filtered using clinically-relevant concepts and their relations. With analysis based on the annotations from the TREC-COVID shared task, we obtain quantitative as well as qualitative insights into characteristics of relational and concept-based literature exploration. Most importantly, we find that the relational concept selection filters the original retrieved collection in a way that decreases the proportion of unjudged documents and increases the precision, which means that the user is likely to be exposed to a larger number of relevant documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge