Antonio Jimeno Yepes
Evolutionary thoughts: integration of large language models and evolutionary algorithms
May 09, 2025Abstract:Large Language Models (LLMs) have unveiled remarkable capabilities in understanding and generating both natural language and code, but LLM reasoning is prone to hallucination and struggle with complex, novel scenarios, often getting stuck on partial or incorrect solutions. However, the inherent ability of Evolutionary Algorithms (EAs) to explore extensive and complex search spaces makes them particularly effective in scenarios where traditional optimization methodologies may falter. However, EAs explore a vast search space when applied to complex problems. To address the computational bottleneck of evaluating large populations, particularly crucial for complex evolutionary tasks, we introduce a highly efficient evaluation framework. This implementation maintains compatibility with existing primitive definitions, ensuring the generation of valid individuals. Using LLMs, we propose an enhanced evolutionary search strategy that enables a more focused exploration of expansive solution spaces. LLMs facilitate the generation of superior candidate solutions, as evidenced by empirical results demonstrating their efficacy in producing improved outcomes.
Financial Report Chunking for Effective Retrieval Augmented Generation
Feb 10, 2024



Abstract:Chunking information is a key step in Retrieval Augmented Generation (RAG). Current research primarily centers on paragraph-level chunking. This approach treats all texts as equal and neglects the information contained in the structure of documents. We propose an expanded approach to chunk documents by moving beyond mere paragraph-level chunking to chunk primary by structural element components of documents. Dissecting documents into these constituent elements creates a new way to chunk documents that yields the best chunk size without tuning. We introduce a novel framework that evaluates how chunking based on element types annotated by document understanding models contributes to the overall context and accuracy of the information retrieved. We also demonstrate how this approach impacts RAG assisted Question & Answer task performance. Our research includes a comprehensive analysis of various element types, their role in effective information retrieval, and the impact they have on the quality of RAG outputs. Findings support that element type based chunking largely improve RAG results on financial reporting. Through this research, we are also able to answer how to uncover highly accurate RAG.
Hypergraph Convolutional Networks for Fine-grained ICU Patient Similarity Analysis and Risk Prediction
Aug 24, 2023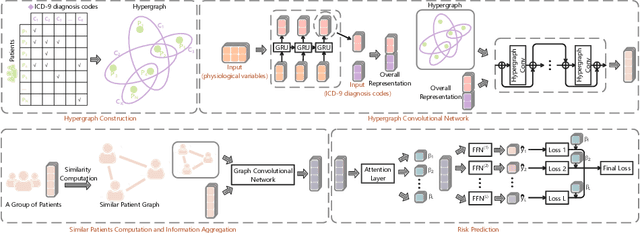
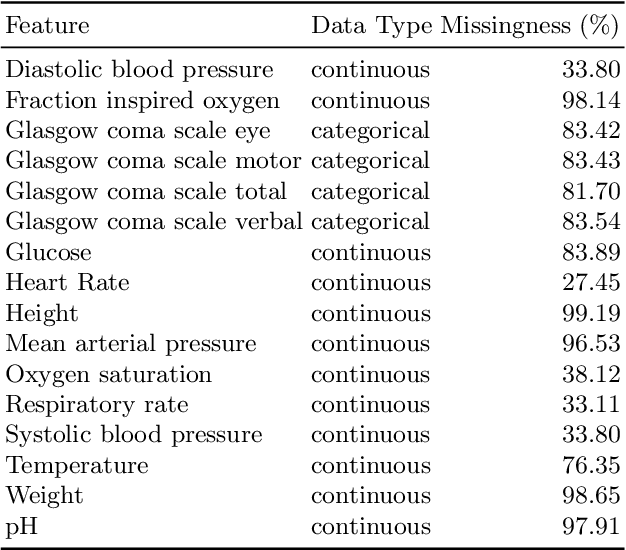
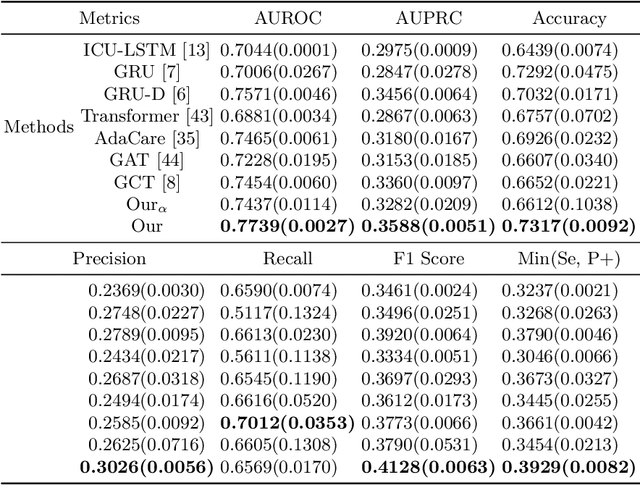
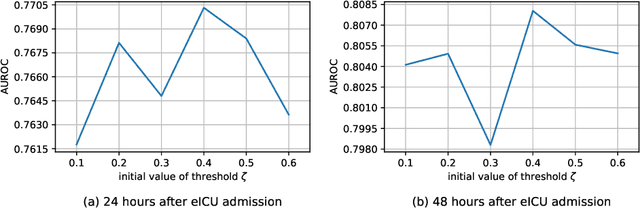
Abstract:The Intensive Care Unit (ICU) is one of the most important parts of a hospital, which admits critically ill patients and provides continuous monitoring and treatment. Various patient outcome prediction methods have been attempted to assist healthcare professionals in clinical decision-making. Existing methods focus on measuring the similarity between patients using deep neural networks to capture the hidden feature structures. However, the higher-order relationships are ignored, such as patient characteristics (e.g., diagnosis codes) and their causal effects on downstream clinical predictions. In this paper, we propose a novel Hypergraph Convolutional Network that allows the representation of non-pairwise relationships among diagnosis codes in a hypergraph to capture the hidden feature structures so that fine-grained patient similarity can be calculated for personalized mortality risk prediction. Evaluation using a publicly available eICU Collaborative Research Database indicates that our method achieves superior performance over the state-of-the-art models on mortality risk prediction. Moreover, the results of several case studies demonstrated the effectiveness of constructing graph networks in providing good transparency and robustness in decision-making.
Contrastive Learning-based Imputation-Prediction Networks for In-hospital Mortality Risk Modeling using EHRs
Aug 19, 2023Abstract:Predicting the risk of in-hospital mortality from electronic health records (EHRs) has received considerable attention. Such predictions will provide early warning of a patient's health condition to healthcare professionals so that timely interventions can be taken. This prediction task is challenging since EHR data are intrinsically irregular, with not only many missing values but also varying time intervals between medical records. Existing approaches focus on exploiting the variable correlations in patient medical records to impute missing values and establishing time-decay mechanisms to deal with such irregularity. This paper presents a novel contrastive learning-based imputation-prediction network for predicting in-hospital mortality risks using EHR data. Our approach introduces graph analysis-based patient stratification modeling in the imputation process to group similar patients. This allows information of similar patients only to be used, in addition to personal contextual information, for missing value imputation. Moreover, our approach can integrate contrastive learning into the proposed network architecture to enhance patient representation learning and predictive performance on the classification task. Experiments on two real-world EHR datasets show that our approach outperforms the state-of-the-art approaches in both imputation and prediction tasks.
Integrated Convolutional and Recurrent Neural Networks for Health Risk Prediction using Patient Journey Data with Many Missing Values
Nov 14, 2022Abstract:Predicting the health risks of patients using Electronic Health Records (EHR) has attracted considerable attention in recent years, especially with the development of deep learning techniques. Health risk refers to the probability of the occurrence of a specific health outcome for a specific patient. The predicted risks can be used to support decision-making by healthcare professionals. EHRs are structured patient journey data. Each patient journey contains a chronological set of clinical events, and within each clinical event, there is a set of clinical/medical activities. Due to variations of patient conditions and treatment needs, EHR patient journey data has an inherently high degree of missingness that contains important information affecting relationships among variables, including time. Existing deep learning-based models generate imputed values for missing values when learning the relationships. However, imputed data in EHR patient journey data may distort the clinical meaning of the original EHR patient journey data, resulting in classification bias. This paper proposes a novel end-to-end approach to modeling EHR patient journey data with Integrated Convolutional and Recurrent Neural Networks. Our model can capture both long- and short-term temporal patterns within each patient journey and effectively handle the high degree of missingness in EHR data without any imputation data generation. Extensive experimental results using the proposed model on two real-world datasets demonstrate robust performance as well as superior prediction accuracy compared to existing state-of-the-art imputation-based prediction methods.
Modeling Long-term Dependencies and Short-term Correlations in Patient Journey Data with Temporal Attention Networks for Health Prediction
Jul 15, 2022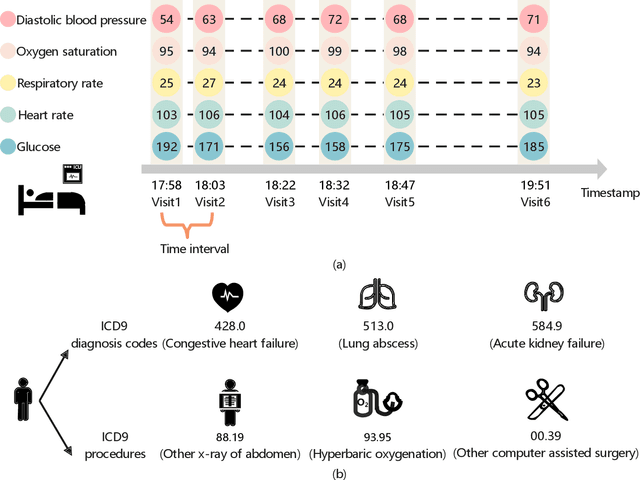
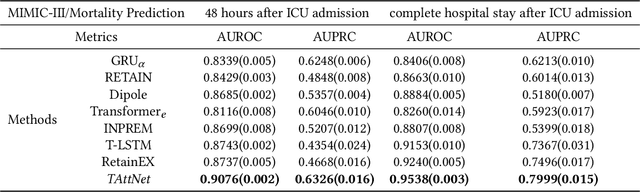
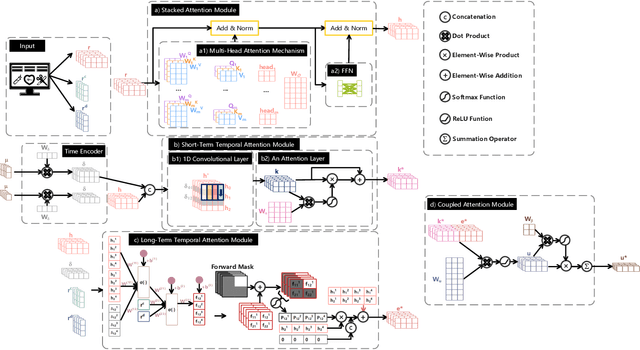

Abstract:Building models for health prediction based on Electronic Health Records (EHR) has become an active research area. EHR patient journey data consists of patient time-ordered clinical events/visits from patients. Most existing studies focus on modeling long-term dependencies between visits, without explicitly taking short-term correlations between consecutive visits into account, where irregular time intervals, incorporated as auxiliary information, are fed into health prediction models to capture latent progressive patterns of patient journeys. We present a novel deep neural network with four modules to take into account the contributions of various variables for health prediction: i) the Stacked Attention module strengthens the deep semantics in clinical events within each patient journey and generates visit embeddings, ii) the Short-Term Temporal Attention module models short-term correlations between consecutive visit embeddings while capturing the impact of time intervals within those visit embeddings, iii) the Long-Term Temporal Attention module models long-term dependencies between visit embeddings while capturing the impact of time intervals within those visit embeddings, iv) and finally, the Coupled Attention module adaptively aggregates the outputs of Short-Term Temporal Attention and Long-Term Temporal Attention modules to make health predictions. Experimental results on MIMIC-III demonstrate superior predictive accuracy of our model compared to existing state-of-the-art methods, as well as the interpretability and robustness of this approach. Furthermore, we found that modeling short-term correlations contributes to local priors generation, leading to improved predictive modeling of patient journeys.
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022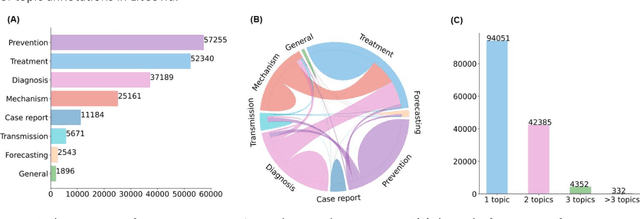
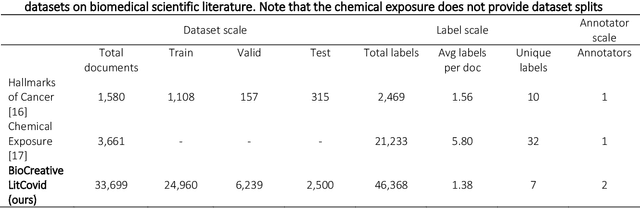
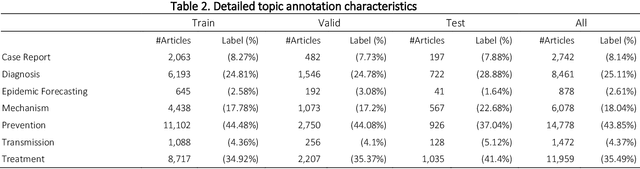
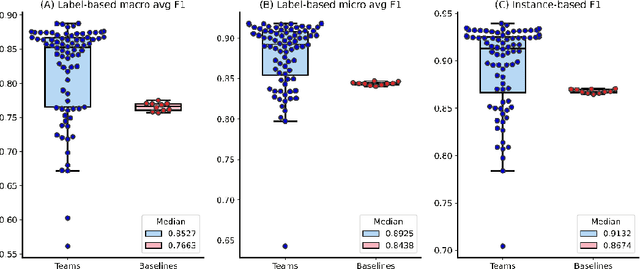
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
Hyperplane bounds for neural feature mappings
Jan 15, 2022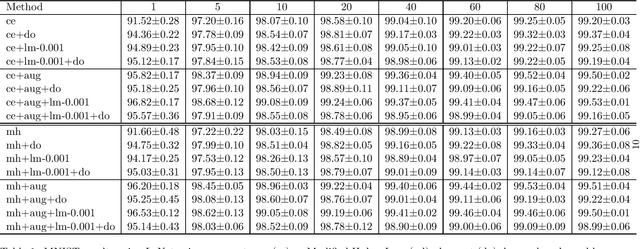

Abstract:Deep learning methods minimise the empirical risk using loss functions such as the cross entropy loss. When minimising the empirical risk, the generalisation of the learnt function still depends on the performance on the training data, the Vapnik-Chervonenkis(VC)-dimension of the function and the number of training examples. Neural networks have a large number of parameters, which correlates with their VC-dimension that is typically large but not infinite, and typically a large number of training instances are needed to effectively train them. In this work, we explore how to optimize feature mappings using neural network with the intention to reduce the effective VC-dimension of the hyperplane found in the space generated by the mapping. An interpretation of the results of this study is that it is possible to define a loss that controls the VC-dimension of the separating hyperplane. We evaluate this approach and observe that the performance when using this method improves when the size of the training set is small.
ICDAR 2021 Competition on Scientific Literature Parsing
Jun 08, 2021
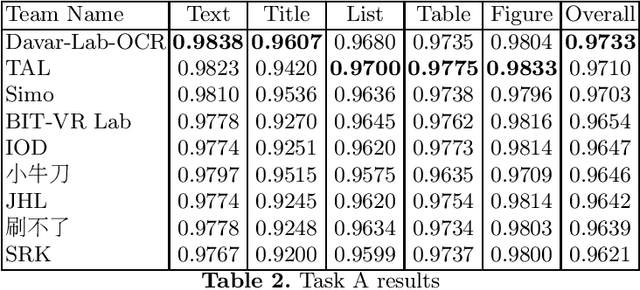
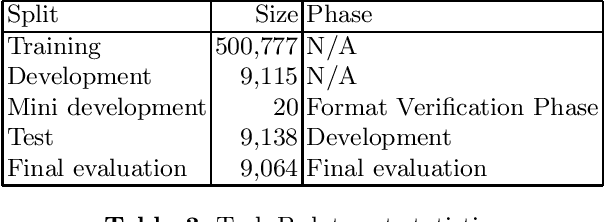
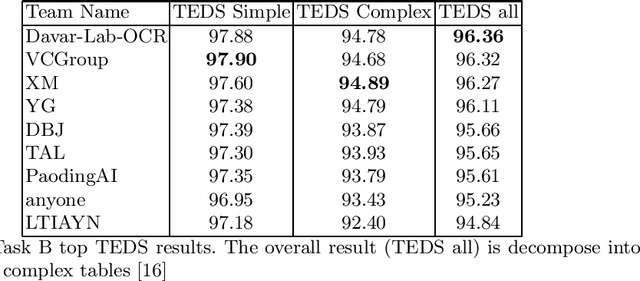
Abstract:Scientific literature contain important information related to cutting-edge innovations in diverse domains. Advances in natural language processing have been driving the fast development in automated information extraction from scientific literature. However, scientific literature is often available in unstructured PDF format. While PDF is great for preserving basic visual elements, such as characters, lines, shapes, etc., on a canvas for presentation to humans, automatic processing of the PDF format by machines presents many challenges. With over 2.5 trillion PDF documents in existence, these issues are prevalent in many other important application domains as well. Our ICDAR 2021 Scientific Literature Parsing Competition (ICDAR2021-SLP) aims to drive the advances specifically in document understanding. ICDAR2021-SLP leverages the PubLayNet and PubTabNet datasets, which provide hundreds of thousands of training and evaluation examples. In Task A, Document Layout Recognition, submissions with the highest performance combine object detection and specialised solutions for the different categories. In Task B, Table Recognition, top submissions rely on methods to identify table components and post-processing methods to generate the table structure and content. Results from both tasks show an impressive performance and opens the possibility for high performance practical applications.
Impact of detecting clinical trial elements in exploration of COVID-19 literature
May 25, 2021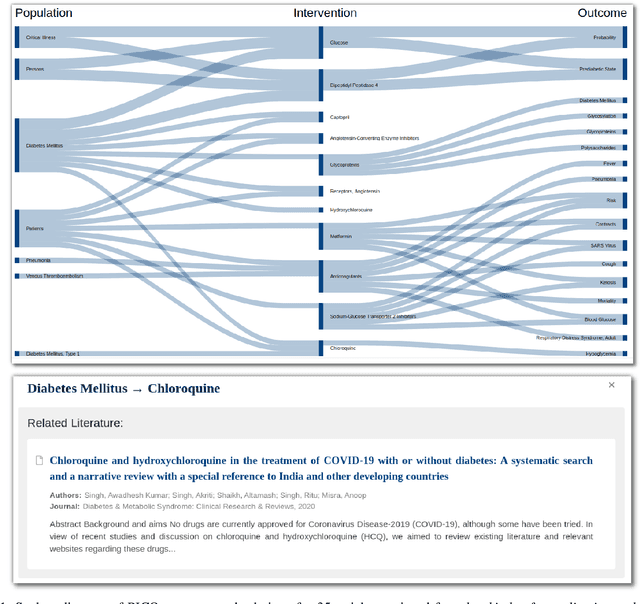
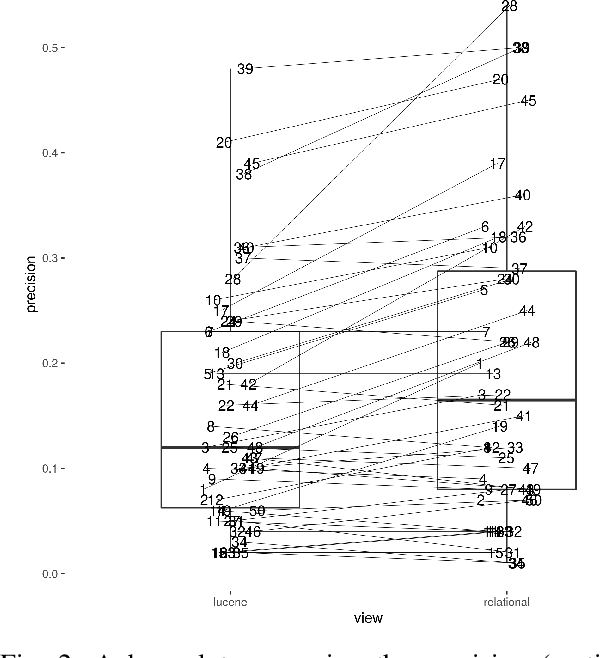
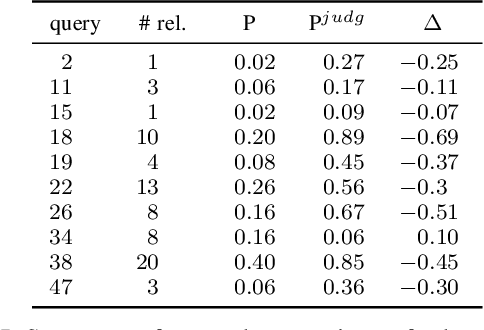

Abstract:The COVID-19 pandemic has driven ever-greater demand for tools which enable efficient exploration of biomedical literature. Although semi-structured information resulting from concept recognition and detection of the defining elements of clinical trials (e.g. PICO criteria) has been commonly used to support literature search, the contributions of this abstraction remain poorly understood, especially in relation to text-based retrieval. In this study, we compare the results retrieved by a standard search engine with those filtered using clinically-relevant concepts and their relations. With analysis based on the annotations from the TREC-COVID shared task, we obtain quantitative as well as qualitative insights into characteristics of relational and concept-based literature exploration. Most importantly, we find that the relational concept selection filters the original retrieved collection in a way that decreases the proportion of unjudged documents and increases the precision, which means that the user is likely to be exposed to a larger number of relevant documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge