Simon Šuster
Impact of detecting clinical trial elements in exploration of COVID-19 literature
May 25, 2021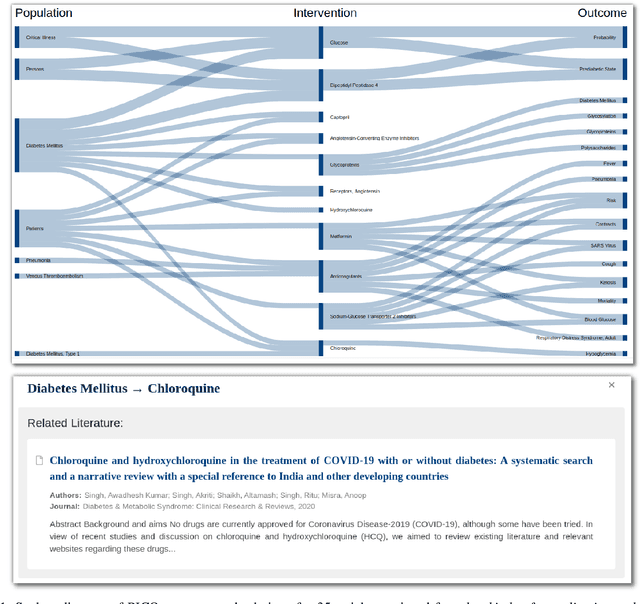
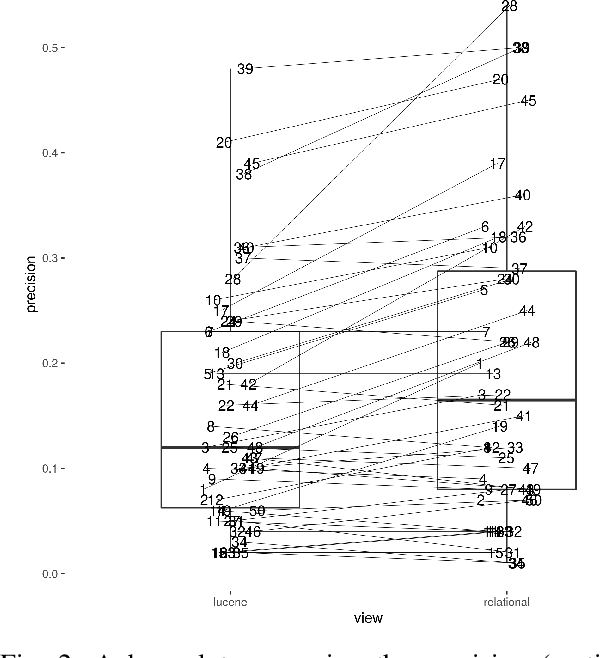
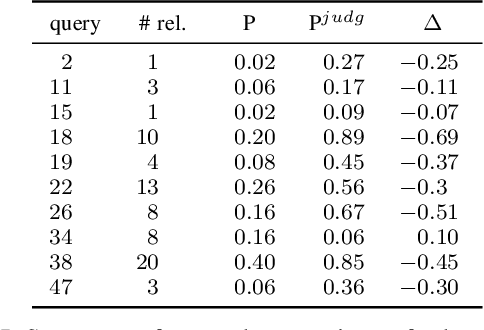

Abstract:The COVID-19 pandemic has driven ever-greater demand for tools which enable efficient exploration of biomedical literature. Although semi-structured information resulting from concept recognition and detection of the defining elements of clinical trials (e.g. PICO criteria) has been commonly used to support literature search, the contributions of this abstraction remain poorly understood, especially in relation to text-based retrieval. In this study, we compare the results retrieved by a standard search engine with those filtered using clinically-relevant concepts and their relations. With analysis based on the annotations from the TREC-COVID shared task, we obtain quantitative as well as qualitative insights into characteristics of relational and concept-based literature exploration. Most importantly, we find that the relational concept selection filters the original retrieved collection in a way that decreases the proportion of unjudged documents and increases the precision, which means that the user is likely to be exposed to a larger number of relevant documents.
COVID-SEE: Scientific Evidence Explorer for COVID-19 Related Research
Aug 18, 2020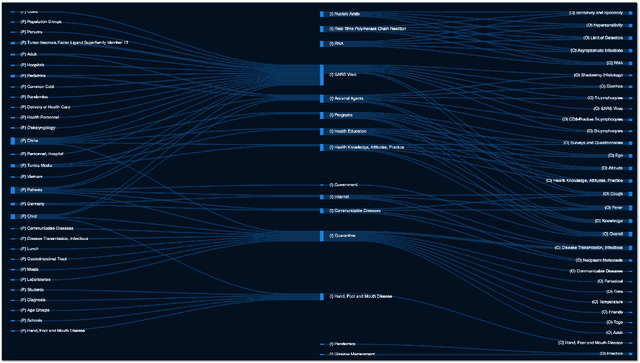
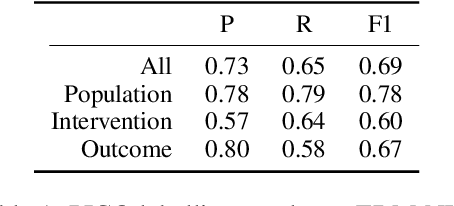
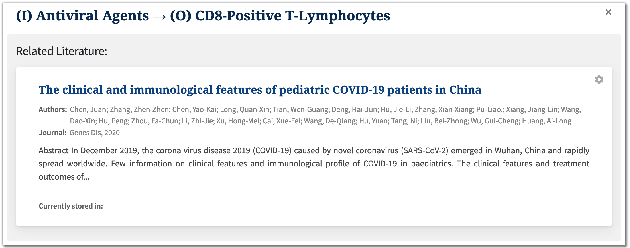
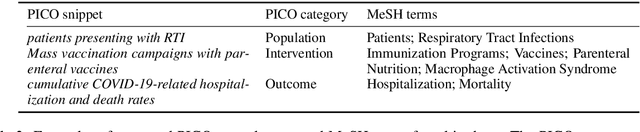
Abstract:We present COVID-SEE, a system for medical literature discovery based on the concept of information exploration, which builds on several distinct text analysis and natural language processing methods to structure and organise information in publications, and augments search by providing a visual overview supporting exploration of a collection to identify key articles of interest. We developed this system over COVID-19 literature to help medical professionals and researchers explore the literature evidence, and improve findability of relevant information. COVID-SEE is available at http://covid-see.com.
Distilling neural networks into skipgram-level decision lists
May 18, 2020



Abstract:Several previous studies on explanation for recurrent neural networks focus on approaches that find the most important input segments for a network as its explanations. In that case, the manner in which these input segments combine with each other to form an explanatory pattern remains unknown. To overcome this, some previous work tries to find patterns (called rules) in the data that explain neural outputs. However, their explanations are often insensitive to model parameters, which limits the scalability of text explanations. To overcome these limitations, we propose a pipeline to explain RNNs by means of decision lists (also called rules) over skipgrams. For evaluation of explanations, we create a synthetic sepsis-identification dataset, as well as apply our technique on additional clinical and sentiment analysis datasets. We find that our technique persistently achieves high explanation fidelity and qualitatively interpretable rules.
Why can't memory networks read effectively?
Oct 16, 2019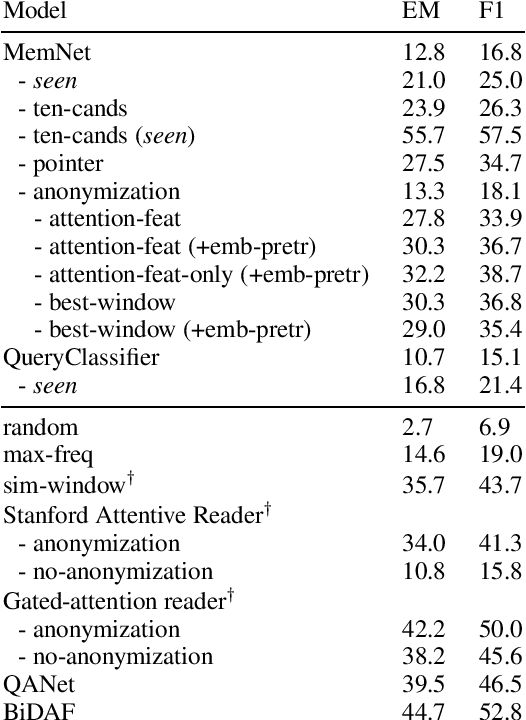
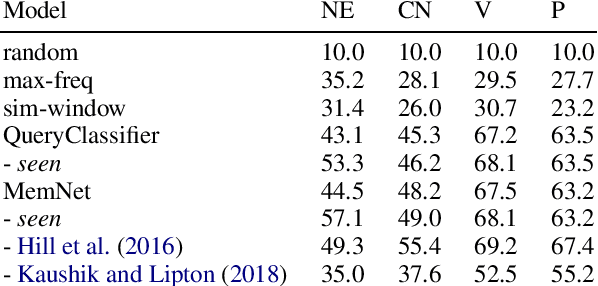

Abstract:Memory networks have been a popular choice among neural architectures for machine reading comprehension and question answering. While recent work revealed that memory networks can't truly perform multi-hop reasoning, we show in the present paper that vanilla memory networks are ineffective even in single-hop reading comprehension. We analyze the reasons for this on two cloze-style datasets, one from the medical domain and another including children's fiction. We find that the output classification layer with entity-specific weights, and the aggregation of passage information with relatively flat attention distributions are the most important contributors to poor results. We propose network adaptations that can serve as simple remedies. We also find that the presence of unseen answers at test time can dramatically affect the reported results, so we suggest controlling for this factor during evaluation.
Rule induction for global explanation of trained models
Aug 29, 2018



Abstract:Understanding the behavior of a trained network and finding explanations for its outputs is important for improving the network's performance and generalization ability, and for ensuring trust in automated systems. Several approaches have previously been proposed to identify and visualize the most important features by analyzing a trained network. However, the relations between different features and classes are lost in most cases. We propose a technique to induce sets of if-then-else rules that capture these relations to globally explain the predictions of a network. We first calculate the importance of the features in the trained network. We then weigh the original inputs with these feature importance scores, simplify the transformed input space, and finally fit a rule induction model to explain the model predictions. We find that the output rule-sets can explain the predictions of a neural network trained for 4-class text classification from the 20 newsgroups dataset to a macro-averaged F-score of 0.80. We make the code available at https://github.com/clips/interpret_with_rules.
Patient representation learning and interpretable evaluation using clinical notes
Jul 03, 2018



Abstract:We have three contributions in this work: 1. We explore the utility of a stacked denoising autoencoder and a paragraph vector model to learn task-independent dense patient representations directly from clinical notes. To analyze if these representations are transferable across tasks, we evaluate them in multiple supervised setups to predict patient mortality, primary diagnostic and procedural category, and gender. We compare their performance with sparse representations obtained from a bag-of-words model. We observe that the learned generalized representations significantly outperform the sparse representations when we have few positive instances to learn from, and there is an absence of strong lexical features. 2. We compare the model performance of the feature set constructed from a bag of words to that obtained from medical concepts. In the latter case, concepts represent problems, treatments, and tests. We find that concept identification does not improve the classification performance. 3. We propose novel techniques to facilitate model interpretability. To understand and interpret the representations, we explore the best encoded features within the patient representations obtained from the autoencoder model. Further, we calculate feature sensitivity across two networks to identify the most significant input features for different classification tasks when we use these pretrained representations as the supervised input. We successfully extract the most influential features for the pipeline using this technique.
* Accepted manuscript at Journal of Biomedical Informatics
CliCR: A Dataset of Clinical Case Reports for Machine Reading Comprehension
Mar 26, 2018



Abstract:We present a new dataset for machine comprehension in the medical domain. Our dataset uses clinical case reports with around 100,000 gap-filling queries about these cases. We apply several baselines and state-of-the-art neural readers to the dataset, and observe a considerable gap in performance (20% F1) between the best human and machine readers. We analyze the skills required for successful answering and show how reader performance varies depending on the applicable skills. We find that inferences using domain knowledge and object tracking are the most frequently required skills, and that recognizing omitted information and spatio-temporal reasoning are the most difficult for the machines.
Unsupervised patient representations from clinical notes with interpretable classification decisions
Nov 14, 2017

Abstract:We have two main contributions in this work: 1. We explore the usage of a stacked denoising autoencoder, and a paragraph vector model to learn task-independent dense patient representations directly from clinical notes. We evaluate these representations by using them as features in multiple supervised setups, and compare their performance with those of sparse representations. 2. To understand and interpret the representations, we explore the best encoded features within the patient representations obtained from the autoencoder model. Further, we calculate the significance of the input features of the trained classifiers when we use these pretrained representations as input.
Unsupervised Context-Sensitive Spelling Correction of English and Dutch Clinical Free-Text with Word and Character N-Gram Embeddings
Oct 19, 2017



Abstract:We present an unsupervised context-sensitive spelling correction method for clinical free-text that uses word and character n-gram embeddings. Our method generates misspelling replacement candidates and ranks them according to their semantic fit, by calculating a weighted cosine similarity between the vectorized representation of a candidate and the misspelling context. To tune the parameters of this model, we generate self-induced spelling error corpora. We perform our experiments for two languages. For English, we greatly outperform off-the-shelf spelling correction tools on a manually annotated MIMIC-III test set, and counter the frequency bias of a noisy channel model, showing that neural embeddings can be successfully exploited to improve upon the state-of-the-art. For Dutch, we also outperform an off-the-shelf spelling correction tool on manually annotated clinical records from the Antwerp University Hospital, but can offer no empirical evidence that our method counters the frequency bias of a noisy channel model in this case as well. However, both our context-sensitive model and our implementation of the noisy channel model obtain high scores on the test set, establishing a state-of-the-art for Dutch clinical spelling correction with the noisy channel model.
* Appears in volume 7 of the CLIN Journal, http://www.clinjournal.org/biblio/volume
A Short Review of Ethical Challenges in Clinical Natural Language Processing
Mar 29, 2017Abstract:Clinical NLP has an immense potential in contributing to how clinical practice will be revolutionized by the advent of large scale processing of clinical records. However, this potential has remained largely untapped due to slow progress primarily caused by strict data access policies for researchers. In this paper, we discuss the concern for privacy and the measures it entails. We also suggest sources of less sensitive data. Finally, we draw attention to biases that can compromise the validity of empirical research and lead to socially harmful applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge