Impact of detecting clinical trial elements in exploration of COVID-19 literature
Paper and Code
May 25, 2021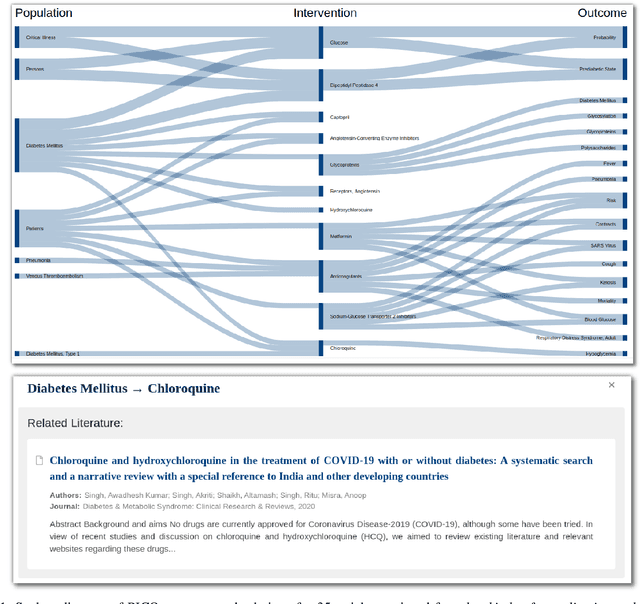
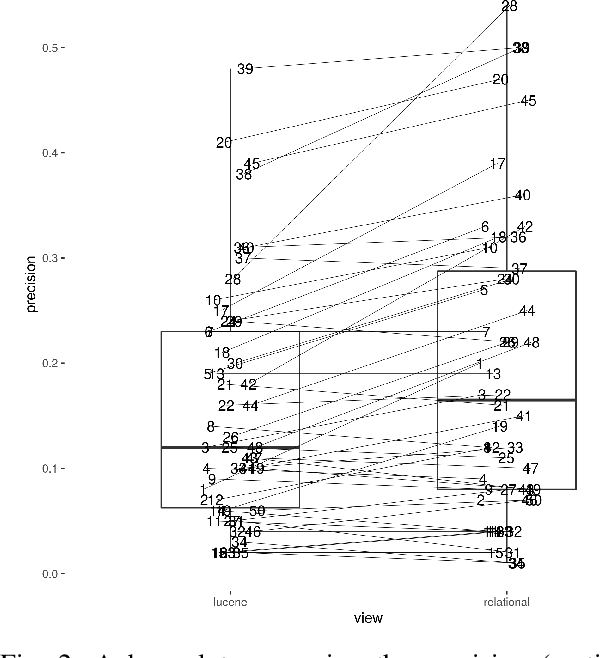
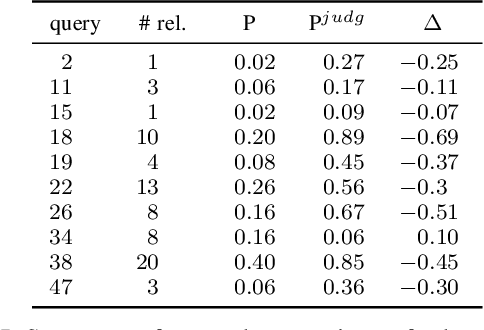

The COVID-19 pandemic has driven ever-greater demand for tools which enable efficient exploration of biomedical literature. Although semi-structured information resulting from concept recognition and detection of the defining elements of clinical trials (e.g. PICO criteria) has been commonly used to support literature search, the contributions of this abstraction remain poorly understood, especially in relation to text-based retrieval. In this study, we compare the results retrieved by a standard search engine with those filtered using clinically-relevant concepts and their relations. With analysis based on the annotations from the TREC-COVID shared task, we obtain quantitative as well as qualitative insights into characteristics of relational and concept-based literature exploration. Most importantly, we find that the relational concept selection filters the original retrieved collection in a way that decreases the proportion of unjudged documents and increases the precision, which means that the user is likely to be exposed to a larger number of relevant documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge