Shaowen Qin
Satisfaction-Aware Incentive Scheme for Federated Learning in Industrial Metaverse: DRL-Based Stackbelberg Game Approach
Feb 10, 2025Abstract:Industrial Metaverse leverages the Industrial Internet of Things (IIoT) to integrate data from diverse devices, employing federated learning and meta-computing to train models in a distributed manner while ensuring data privacy. Achieving an immersive experience for industrial Metaverse necessitates maintaining a balance between model quality and training latency. Consequently, a primary challenge in federated learning tasks is optimizing overall system performance by balancing model quality and training latency. This paper designs a satisfaction function that accounts for data size, Age of Information (AoI), and training latency. Additionally, the satisfaction function is incorporated into the utility functions to incentivize node participation in model training. We model the utility functions of servers and nodes as a two-stage Stackelberg game and employ a deep reinforcement learning approach to learn the Stackelberg equilibrium. This approach ensures balanced rewards and enhances the applicability of the incentive scheme for industrial Metaverse. Simulation results demonstrate that, under the same budget constraints, the proposed incentive scheme improves at least 23.7% utility compared to existing schemes without compromising model accuracy.
Hypergraph Convolutional Networks for Fine-grained ICU Patient Similarity Analysis and Risk Prediction
Aug 24, 2023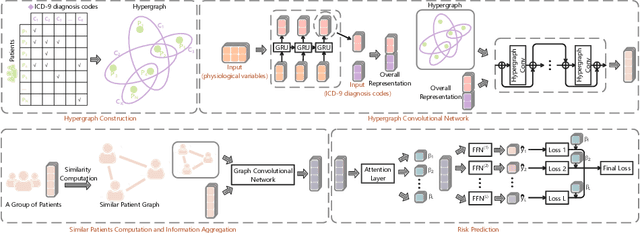
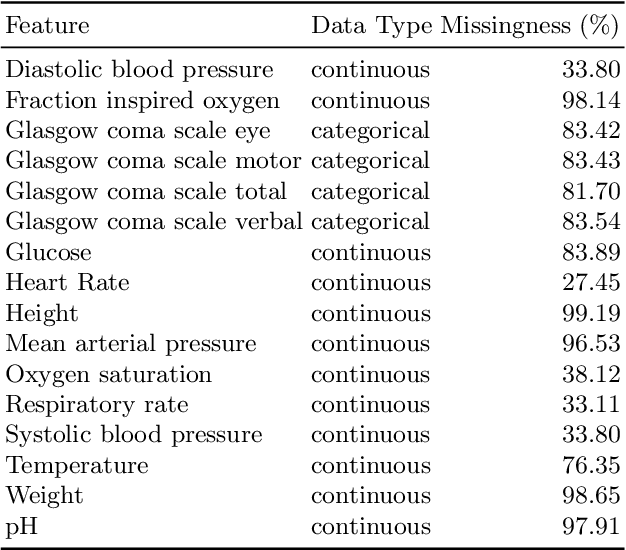
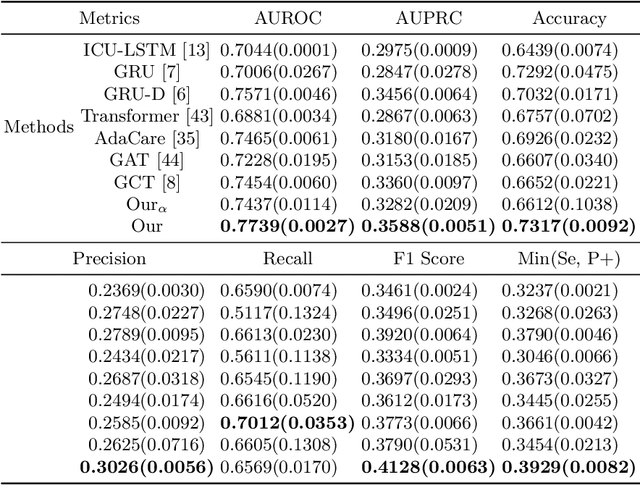
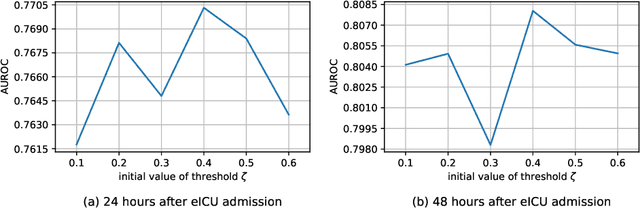
Abstract:The Intensive Care Unit (ICU) is one of the most important parts of a hospital, which admits critically ill patients and provides continuous monitoring and treatment. Various patient outcome prediction methods have been attempted to assist healthcare professionals in clinical decision-making. Existing methods focus on measuring the similarity between patients using deep neural networks to capture the hidden feature structures. However, the higher-order relationships are ignored, such as patient characteristics (e.g., diagnosis codes) and their causal effects on downstream clinical predictions. In this paper, we propose a novel Hypergraph Convolutional Network that allows the representation of non-pairwise relationships among diagnosis codes in a hypergraph to capture the hidden feature structures so that fine-grained patient similarity can be calculated for personalized mortality risk prediction. Evaluation using a publicly available eICU Collaborative Research Database indicates that our method achieves superior performance over the state-of-the-art models on mortality risk prediction. Moreover, the results of several case studies demonstrated the effectiveness of constructing graph networks in providing good transparency and robustness in decision-making.
Contrastive Learning-based Imputation-Prediction Networks for In-hospital Mortality Risk Modeling using EHRs
Aug 19, 2023Abstract:Predicting the risk of in-hospital mortality from electronic health records (EHRs) has received considerable attention. Such predictions will provide early warning of a patient's health condition to healthcare professionals so that timely interventions can be taken. This prediction task is challenging since EHR data are intrinsically irregular, with not only many missing values but also varying time intervals between medical records. Existing approaches focus on exploiting the variable correlations in patient medical records to impute missing values and establishing time-decay mechanisms to deal with such irregularity. This paper presents a novel contrastive learning-based imputation-prediction network for predicting in-hospital mortality risks using EHR data. Our approach introduces graph analysis-based patient stratification modeling in the imputation process to group similar patients. This allows information of similar patients only to be used, in addition to personal contextual information, for missing value imputation. Moreover, our approach can integrate contrastive learning into the proposed network architecture to enhance patient representation learning and predictive performance on the classification task. Experiments on two real-world EHR datasets show that our approach outperforms the state-of-the-art approaches in both imputation and prediction tasks.
Integrated Convolutional and Recurrent Neural Networks for Health Risk Prediction using Patient Journey Data with Many Missing Values
Nov 14, 2022Abstract:Predicting the health risks of patients using Electronic Health Records (EHR) has attracted considerable attention in recent years, especially with the development of deep learning techniques. Health risk refers to the probability of the occurrence of a specific health outcome for a specific patient. The predicted risks can be used to support decision-making by healthcare professionals. EHRs are structured patient journey data. Each patient journey contains a chronological set of clinical events, and within each clinical event, there is a set of clinical/medical activities. Due to variations of patient conditions and treatment needs, EHR patient journey data has an inherently high degree of missingness that contains important information affecting relationships among variables, including time. Existing deep learning-based models generate imputed values for missing values when learning the relationships. However, imputed data in EHR patient journey data may distort the clinical meaning of the original EHR patient journey data, resulting in classification bias. This paper proposes a novel end-to-end approach to modeling EHR patient journey data with Integrated Convolutional and Recurrent Neural Networks. Our model can capture both long- and short-term temporal patterns within each patient journey and effectively handle the high degree of missingness in EHR data without any imputation data generation. Extensive experimental results using the proposed model on two real-world datasets demonstrate robust performance as well as superior prediction accuracy compared to existing state-of-the-art imputation-based prediction methods.
Compound Density Networks for Risk Prediction using Electronic Health Records
Aug 16, 2022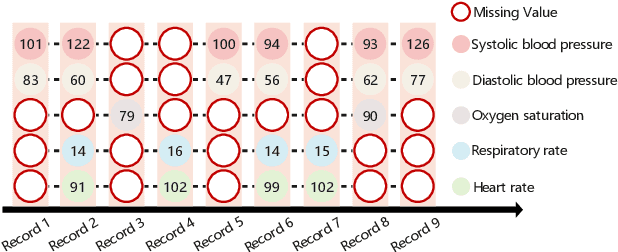
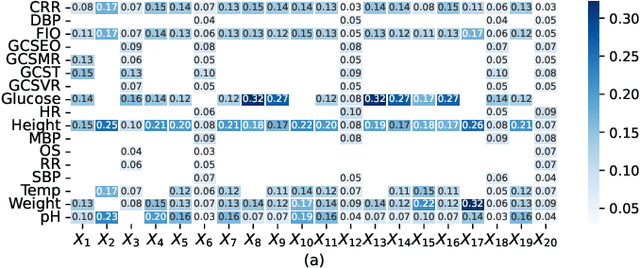
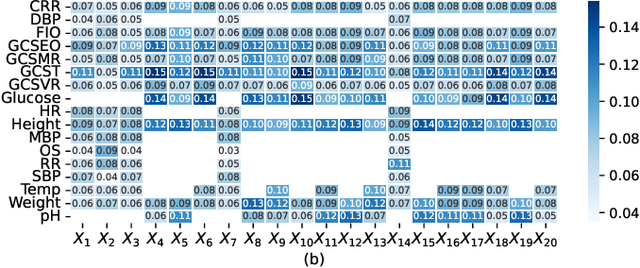
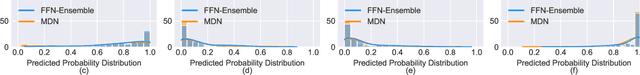
Abstract:Electronic Health Records (EHRs) exhibit a high amount of missing data due to variations of patient conditions and treatment needs. Imputation of missing values has been considered an effective approach to deal with this challenge. Existing work separates imputation method and prediction model as two independent parts of an EHR-based machine learning system. We propose an integrated end-to-end approach by utilizing a Compound Density Network (CDNet) that allows the imputation method and prediction model to be tuned together within a single framework. CDNet consists of a Gated recurrent unit (GRU), a Mixture Density Network (MDN), and a Regularized Attention Network (RAN). The GRU is used as a latent variable model to model EHR data. The MDN is designed to sample latent variables generated by GRU. The RAN serves as a regularizer for less reliable imputed values. The architecture of CDNet enables GRU and MDN to iteratively leverage the output of each other to impute missing values, leading to a more accurate and robust prediction. We validate CDNet on the mortality prediction task on the MIMIC-III dataset. Our model outperforms state-of-the-art models by significant margins. We also empirically show that regularizing imputed values is a key factor for superior prediction performance. Analysis of prediction uncertainty shows that our model can capture both aleatoric and epistemic uncertainties, which offers model users a better understanding of the model results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge