Yuxi Liu
Aggregation Queries over Unstructured Text: Benchmark and Agentic Method
Feb 03, 2026Abstract:Aggregation query over free text is a long-standing yet underexplored problem. Unlike ordinary question answering, aggregate queries require exhaustive evidence collection and systems are required to "find all," not merely "find one." Existing paradigms such as Text-to-SQL and Retrieval-Augmented Generation fail to achieve this completeness. In this work, we formalize entity-level aggregation querying over text in a corpus-bounded setting with strict completeness requirement. To enable principled evaluation, we introduce AGGBench, a benchmark designed to evaluate completeness-oriented aggregation under realistic large-scale corpus. To accompany the benchmark, we propose DFA (Disambiguation--Filtering--Aggregation), a modular agentic baseline that decomposes aggregation querying into interpretable stages and exposes key failure modes related to ambiguity, filtering, and aggregation. Empirical results show that DFA consistently improves aggregation evidence coverage over strong RAG and agentic baselines. The data and code are available in \href{https://anonymous.4open.science/r/DFA-A4C1}.
DSFedMed: Dual-Scale Federated Medical Image Segmentation via Mutual Distillation Between Foundation and Lightweight Models
Jan 22, 2026Abstract:Foundation Models (FMs) have demonstrated strong generalization across diverse vision tasks. However, their deployment in federated settings is hindered by high computational demands, substantial communication overhead, and significant inference costs. We propose DSFedMed, a dual-scale federated framework that enables mutual knowledge distillation between a centralized foundation model and lightweight client models for medical image segmentation. To support knowledge distillation, a set of high-quality medical images is generated to replace real public datasets, and a learnability-guided sample selection strategy is proposed to enhance efficiency and effectiveness in dual-scale distillation. This mutual distillation enables the foundation model to transfer general knowledge to lightweight clients, while also incorporating client-specific insights to refine the foundation model. Evaluations on five medical imaging segmentation datasets show that DSFedMed achieves an average 2 percent improvement in Dice score while reducing communication costs and inference time by nearly 90 percent compared to existing federated foundation model baselines. These results demonstrate significant efficiency gains and scalability for resource-limited federated deployments.
Mixture of Distributions Matters: Dynamic Sparse Attention for Efficient Video Diffusion Transformers
Jan 14, 2026Abstract:While Diffusion Transformers (DiTs) have achieved notable progress in video generation, this long-sequence generation task remains constrained by the quadratic complexity inherent to self-attention mechanisms, creating significant barriers to practical deployment. Although sparse attention methods attempt to address this challenge, existing approaches either rely on oversimplified static patterns or require computationally expensive sampling operations to achieve dynamic sparsity, resulting in inaccurate pattern predictions and degraded generation quality. To overcome these limitations, we propose a \underline{\textbf{M}}ixtrue-\underline{\textbf{O}}f-\underline{\textbf{D}}istribution \textbf{DiT} (\textbf{MOD-DiT}), a novel sampling-free dynamic attention framework that accurately models evolving attention patterns through a two-stage process. First, MOD-DiT leverages prior information from early denoising steps and adopts a {distributed mixing approach} to model an efficient linear approximation model, which is then used to predict mask patterns for a specific denoising interval. Second, an online block masking strategy dynamically applies these predicted masks while maintaining historical sparsity information, eliminating the need for repetitive sampling operations. Extensive evaluations demonstrate consistent acceleration and quality improvements across multiple benchmarks and model architectures, validating MOD-DiT's effectiveness for efficient, high-quality video generation while overcoming the computational limitations of traditional sparse attention approaches.
Neural B-frame Video Compression with Bi-directional Reference Harmonization
Nov 12, 2025Abstract:Neural video compression (NVC) has made significant progress in recent years, while neural B-frame video compression (NBVC) remains underexplored compared to P-frame compression. NBVC can adopt bi-directional reference frames for better compression performance. However, NBVC's hierarchical coding may complicate continuous temporal prediction, especially at some hierarchical levels with a large frame span, which could cause the contribution of the two reference frames to be unbalanced. To optimize reference information utilization, we propose a novel NBVC method, termed Bi-directional Reference Harmonization Video Compression (BRHVC), with the proposed Bi-directional Motion Converge (BMC) and Bi-directional Contextual Fusion (BCF). BMC converges multiple optical flows in motion compression, leading to more accurate motion compensation on a larger scale. Then BCF explicitly models the weights of reference contexts under the guidance of motion compensation accuracy. With more efficient motions and contexts, BRHVC can effectively harmonize bi-directional references. Experimental results indicate that our BRHVC outperforms previous state-of-the-art NVC methods, even surpassing the traditional coding, VTM-RA (under random access configuration), on the HEVC datasets. The source code is released at https://github.com/kwai/NVC.
Unsupervised Active Learning via Natural Feature Progressive Framework
Oct 06, 2025



Abstract:The effectiveness of modern deep learning models is predicated on the availability of large-scale, human-annotated datasets, a process that is notoriously expensive and time-consuming. While Active Learning (AL) offers a strategic solution by labeling only the most informative and representative data, its iterative nature still necessitates significant human involvement. Unsupervised Active Learning (UAL) presents an alternative by shifting the annotation burden to a single, post-selection step. Unfortunately, prevailing UAL methods struggle to achieve state-of-the-art performance. These approaches typically rely on local, gradient-based scoring for sample importance estimation, which not only makes them vulnerable to ambiguous and noisy data but also hinders their capacity to select samples that adequately represent the full data distribution. Moreover, their use of shallow, one-shot linear selection falls short of a true UAL paradigm. In this paper, we propose the Natural Feature Progressive Framework (NFPF), a UAL method that revolutionizes how sample importance is measured. At its core, NFPF employs a Specific Feature Learning Machine (SFLM) to effectively quantify each sample's contribution to model performance. We further utilize the SFLM to define a powerful Reconstruction Difference metric for initial sample selection. Our comprehensive experiments show that NFPF significantly outperforms all established UAL methods and achieves performance on par with supervised AL methods on vision datasets. Detailed ablation studies and qualitative visualizations provide compelling evidence for NFPF's superior performance, enhanced robustness, and improved data distribution coverage.
Once-for-All: Controllable Generative Image Compression with Dynamic Granularity Adaption
Jun 02, 2024



Abstract:Although recent generative image compression methods have demonstrated impressive potential in optimizing the rate-distortion-perception trade-off, they still face the critical challenge of flexible rate adaption to diverse compression necessities and scenarios. To overcome this challenge, this paper proposes a Controllable Generative Image Compression framework, Control-GIC, the first capable of fine-grained bitrate adaption across a broad spectrum while ensuring high-fidelity and generality compression. We base Control-GIC on a VQGAN framework representing an image as a sequence of variable-length codes (i.e. VQ-indices), which can be losslessly compressed and exhibits a direct positive correlation with the bitrates. Therefore, drawing inspiration from the classical coding principle, we naturally correlate the information density of local image patches with their granular representations, to achieve dynamic adjustment of the code quantity following different granularity decisions. This implies we can flexibly determine a proper allocation of granularity for the patches to acquire desirable compression rates. We further develop a probabilistic conditional decoder that can trace back to historic encoded multi-granularity representations according to transmitted codes, and then reconstruct hierarchical granular features in the formalization of conditional probability, enabling more informative aggregation to improve reconstruction realism. Our experiments show that Control-GIC allows highly flexible and controllable bitrate adaption and even once compression on an entire dataset to fulfill constrained bitrate conditions. Experimental results demonstrate its superior performance over recent state-of-the-art methods.
SelfGNN: Self-Supervised Graph Neural Networks for Sequential Recommendation
May 31, 2024



Abstract:Sequential recommendation effectively addresses information overload by modeling users' temporal and sequential interaction patterns. To overcome the limitations of supervision signals, recent approaches have adopted self-supervised learning techniques in recommender systems. However, there are still two critical challenges that remain unsolved. Firstly, existing sequential models primarily focus on long-term modeling of individual interaction sequences, overlooking the valuable short-term collaborative relationships among the behaviors of different users. Secondly, real-world data often contain noise, particularly in users' short-term behaviors, which can arise from temporary intents or misclicks. Such noise negatively impacts the accuracy of both graph and sequence models, further complicating the modeling process. To address these challenges, we propose a novel framework called Self-Supervised Graph Neural Network (SelfGNN) for sequential recommendation. The SelfGNN framework encodes short-term graphs based on time intervals and utilizes Graph Neural Networks (GNNs) to learn short-term collaborative relationships. It captures long-term user and item representations at multiple granularity levels through interval fusion and dynamic behavior modeling. Importantly, our personalized self-augmented learning structure enhances model robustness by mitigating noise in short-term graphs based on long-term user interests and personal stability. Extensive experiments conducted on four real-world datasets demonstrate that SelfGNN outperforms various state-of-the-art baselines. Our model implementation codes are available at https://github.com/HKUDS/SelfGNN.
FedFMS: Exploring Federated Foundation Models for Medical Image Segmentation
Mar 08, 2024
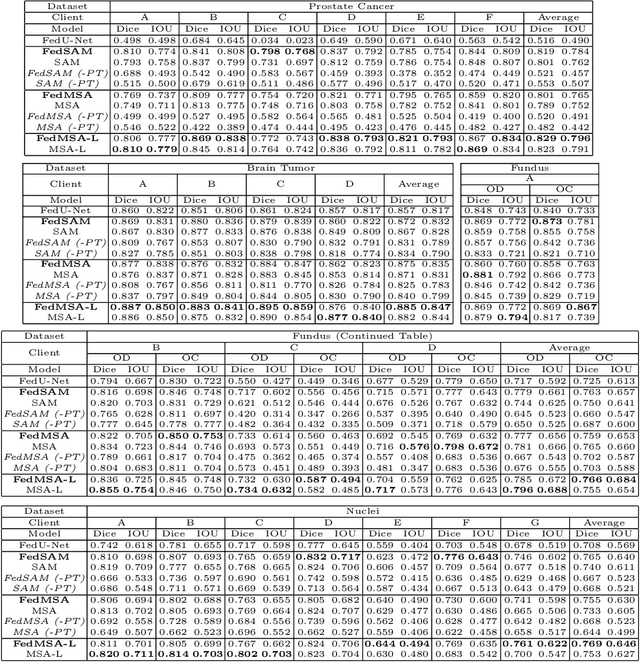


Abstract:Medical image segmentation is crucial for clinical diagnosis. The Segmentation Anything Model (SAM) serves as a powerful foundation model for visual segmentation and can be adapted for medical image segmentation. However, medical imaging data typically contain privacy-sensitive information, making it challenging to train foundation models with centralized storage and sharing. To date, there are few foundation models tailored for medical image deployment within the federated learning framework, and the segmentation performance, as well as the efficiency of communication and training, remain unexplored. In response to these issues, we developed Federated Foundation models for Medical image Segmentation (FedFMS), which includes the Federated SAM (FedSAM) and a communication and training-efficient Federated SAM with Medical SAM Adapter (FedMSA). Comprehensive experiments on diverse datasets are conducted to investigate the performance disparities between centralized training and federated learning across various configurations of FedFMS. The experiments revealed that FedFMS could achieve performance comparable to models trained via centralized training methods while maintaining privacy. Furthermore, FedMSA demonstrated the potential to enhance communication and training efficiency. Our model implementation codes are available at https://github.com/LIU-YUXI/FedFMS.
Region-Adaptive Transform with Segmentation Prior for Image Compression
Mar 01, 2024



Abstract:Learned Image Compression (LIC) has shown remarkable progress in recent years. Existing works commonly employ CNN-based or self-attention-based modules as transform methods for compression. However, there is no prior research on neural transform that focuses on specific regions. In response, we introduce the class-agnostic segmentation masks (i.e. semantic masks without category labels) for extracting region-adaptive contextual information. Our proposed module, Region-Adaptive Transform, applies adaptive convolutions on different regions guided by the masks. Additionally, we introduce a plug-and-play module named Scale Affine Layer to incorporate rich contexts from various regions. While there have been prior image compression efforts that involve segmentation masks as additional intermediate inputs, our approach differs significantly from them. Our advantages lie in that, to avoid extra bitrate overhead, we treat these masks as privilege information, which is accessible during the model training stage but not required during the inference phase. To the best of our knowledge, we are the first to employ class-agnostic masks as privilege information and achieve superior performance in pixel-fidelity metrics, such as Peak Signal to Noise Ratio (PSNR). The experimental results demonstrate our improvement compared to previously well-performing methods, with about 8.2% bitrate saving compared to VTM-17.0. The code will be released at https://github.com/GityuxiLiu/Region-Adaptive-Transform-with-Segmentation-Prior-for-Image-Compression.
Hypergraph Convolutional Networks for Fine-grained ICU Patient Similarity Analysis and Risk Prediction
Aug 24, 2023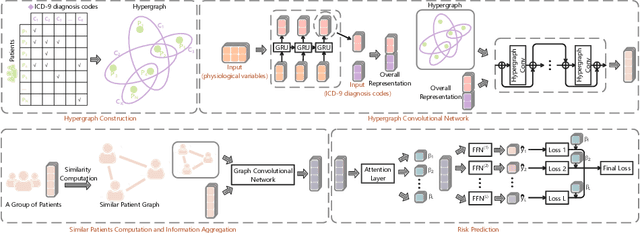
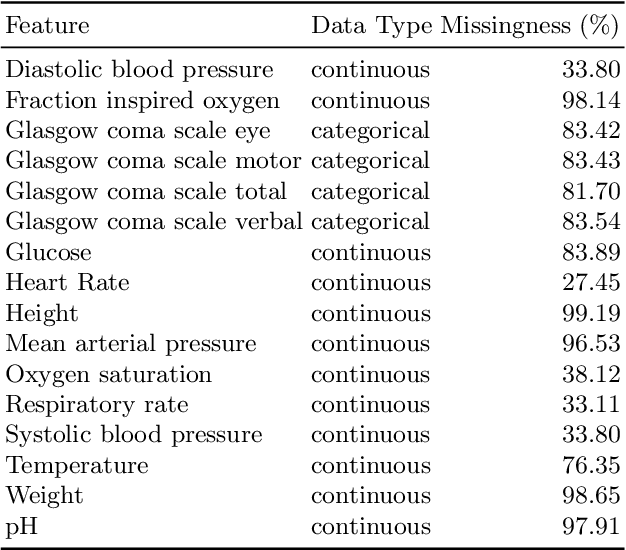
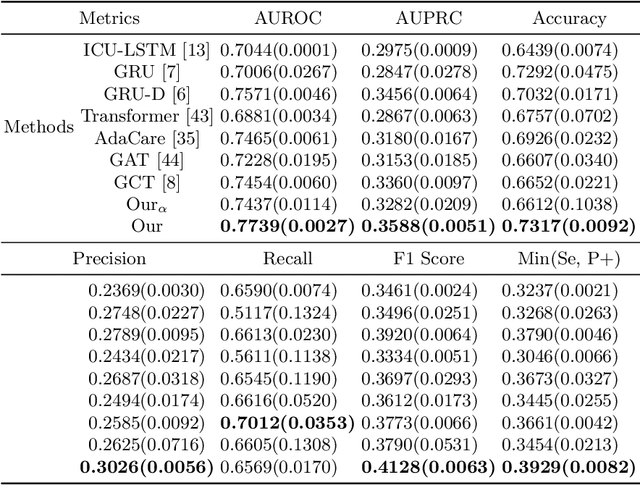
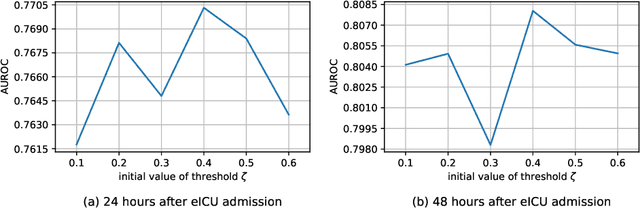
Abstract:The Intensive Care Unit (ICU) is one of the most important parts of a hospital, which admits critically ill patients and provides continuous monitoring and treatment. Various patient outcome prediction methods have been attempted to assist healthcare professionals in clinical decision-making. Existing methods focus on measuring the similarity between patients using deep neural networks to capture the hidden feature structures. However, the higher-order relationships are ignored, such as patient characteristics (e.g., diagnosis codes) and their causal effects on downstream clinical predictions. In this paper, we propose a novel Hypergraph Convolutional Network that allows the representation of non-pairwise relationships among diagnosis codes in a hypergraph to capture the hidden feature structures so that fine-grained patient similarity can be calculated for personalized mortality risk prediction. Evaluation using a publicly available eICU Collaborative Research Database indicates that our method achieves superior performance over the state-of-the-art models on mortality risk prediction. Moreover, the results of several case studies demonstrated the effectiveness of constructing graph networks in providing good transparency and robustness in decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge