Yang Xie
College of Informatics, Huazhong Agricultural University, Wuhan, China
MEDVISTAGYM: A Scalable Training Environment for Thinking with Medical Images via Tool-Integrated Reinforcement Learning
Jan 12, 2026Abstract:Vision language models (VLMs) achieve strong performance on general image understanding but struggle to think with medical images, especially when performing multi-step reasoning through iterative visual interaction. Medical VLMs often rely on static visual embeddings and single-pass inference, preventing models from re-examining, verifying, or refining visual evidence during reasoning. While tool-integrated reasoning offers a promising path forward, open-source VLMs lack the training infrastructure to learn effective tool selection, invocation, and coordination in multi-modal medical reasoning. We introduce MedVistaGym, a scalable and interactive training environment that incentivizes tool-integrated visual reasoning for medical image analysis. MedVistaGym equips VLMs to determine when and which tools to invoke, localize task-relevant image regions, and integrate single or multiple sub-image evidence into interleaved multimodal reasoning within a unified, executable interface for agentic training. Using MedVistaGym, we train MedVistaGym-R1 to interleave tool use with agentic reasoning through trajectory sampling and end-to-end reinforcement learning. Across six medical VQA benchmarks, MedVistaGym-R1-8B exceeds comparably sized tool-augmented baselines by 19.10% to 24.21%, demonstrating that structured agentic training--not tool access alone--unlocks effective tool-integrated reasoning for medical image analysis.
MedAgentGym: Training LLM Agents for Code-Based Medical Reasoning at Scale
Jun 04, 2025



Abstract:We introduce MedAgentGYM, the first publicly available training environment designed to enhance coding-based medical reasoning capabilities in large language model (LLM) agents. MedAgentGYM comprises 72,413 task instances across 129 categories derived from authentic real-world biomedical scenarios. Tasks are encapsulated within executable coding environments, each featuring detailed task descriptions, interactive feedback mechanisms, verifiable ground-truth annotations, and scalable training trajectory generation. Extensive benchmarking of over 30 LLMs reveals a notable performance disparity between commercial API-based models and open-source counterparts. Leveraging MedAgentGYM, Med-Copilot-7B achieves substantial performance gains through supervised fine-tuning (+36.44%) and continued reinforcement learning (+42.47%), emerging as an affordable and privacy-preserving alternative competitive with gpt-4o. By offering both a comprehensive benchmark and accessible, expandable training resources within unified execution environments, MedAgentGYM delivers an integrated platform to develop LLM-based coding assistants for advanced biomedical research and practice.
Large language models enabled multiagent ensemble method for efficient EHR data labeling
Oct 21, 2024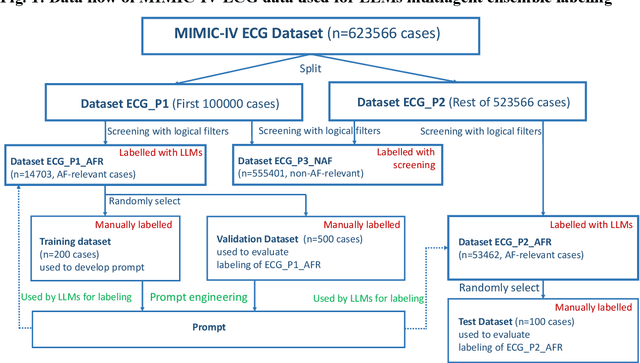
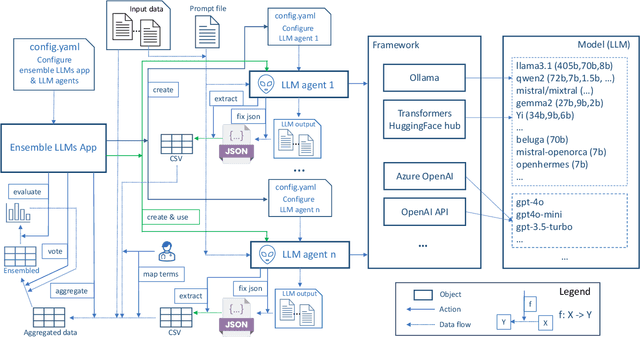
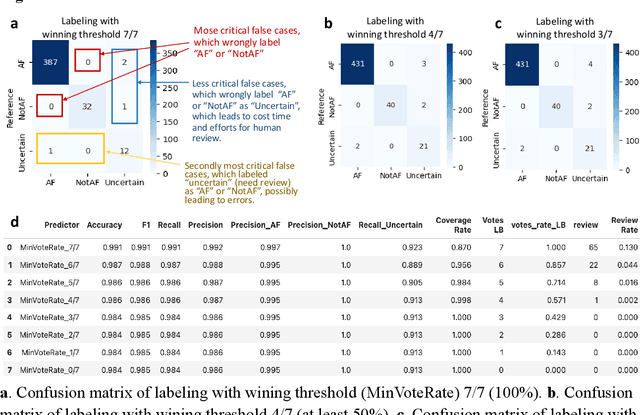
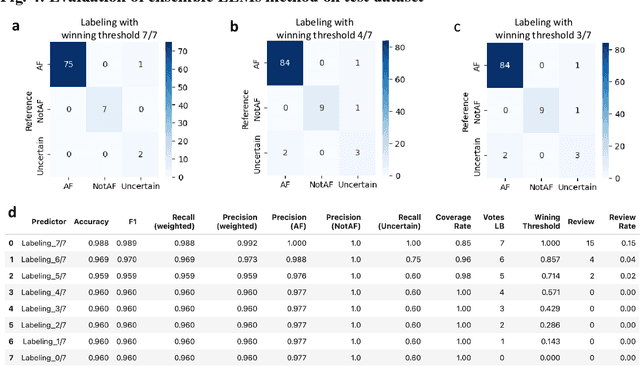
Abstract:This study introduces a novel multiagent ensemble method powered by LLMs to address a key challenge in ML - data labeling, particularly in large-scale EHR datasets. Manual labeling of such datasets requires domain expertise and is labor-intensive, time-consuming, expensive, and error-prone. To overcome this bottleneck, we developed an ensemble LLMs method and demonstrated its effectiveness in two real-world tasks: (1) labeling a large-scale unlabeled ECG dataset in MIMIC-IV; (2) identifying social determinants of health (SDOH) from the clinical notes of EHR. Trading off benefits and cost, we selected a pool of diverse open source LLMs with satisfactory performance. We treat each LLM's prediction as a vote and apply a mechanism of majority voting with minimal winning threshold for ensemble. We implemented an ensemble LLMs application for EHR data labeling tasks. By using the ensemble LLMs and natural language processing, we labeled MIMIC-IV ECG dataset of 623,566 ECG reports with an estimated accuracy of 98.2%. We applied the ensemble LLMs method to identify SDOH from social history sections of 1,405 EHR clinical notes, also achieving competitive performance. Our experiments show that the ensemble LLMs can outperform individual LLM even the best commercial one, and the method reduces hallucination errors. From the research, we found that (1) the ensemble LLMs method significantly reduces the time and effort required for labeling large-scale EHR data, automating the process with high accuracy and quality; (2) the method generalizes well to other text data labeling tasks, as shown by its application to SDOH identification; (3) the ensemble of a group of diverse LLMs can outperform or match the performance of the best individual LLM; and (4) the ensemble method substantially reduces hallucination errors. This approach provides a scalable and efficient solution to data-labeling challenges.
Causal Effect Estimation using identifiable Variational AutoEncoder with Latent Confounders and Post-Treatment Variables
Aug 13, 2024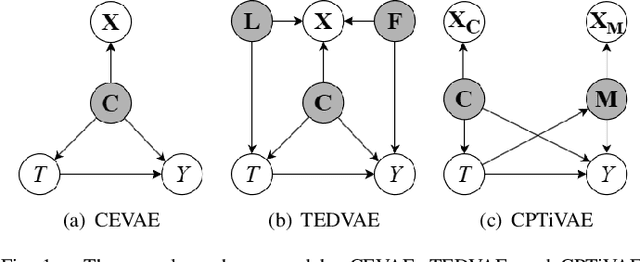
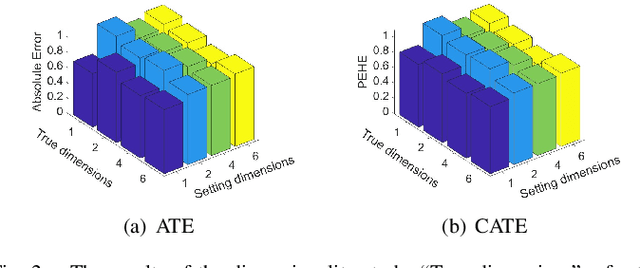
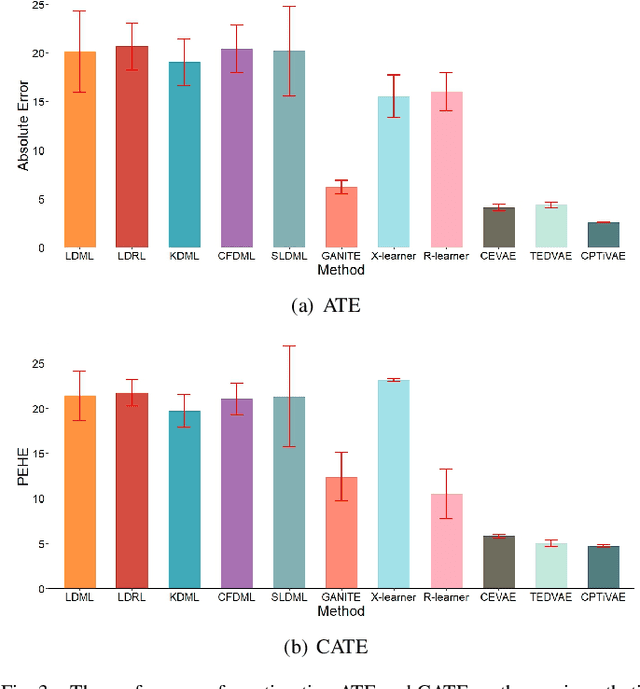
Abstract:Estimating causal effects from observational data is challenging, especially in the presence of latent confounders. Much work has been done on addressing this challenge, but most of the existing research ignores the bias introduced by the post-treatment variables. In this paper, we propose a novel method of joint Variational AutoEncoder (VAE) and identifiable Variational AutoEncoder (iVAE) for learning the representations of latent confounders and latent post-treatment variables from their proxy variables, termed CPTiVAE, to achieve unbiased causal effect estimation from observational data. We further prove the identifiability in terms of the representation of latent post-treatment variables. Extensive experiments on synthetic and semi-synthetic datasets demonstrate that the CPTiVAE outperforms the state-of-the-art methods in the presence of latent confounders and post-treatment variables. We further apply CPTiVAE to a real-world dataset to show its potential application.
Disentangled Latent Representation Learning for Tackling the Confounding M-Bias Problem in Causal Inference
Dec 08, 2023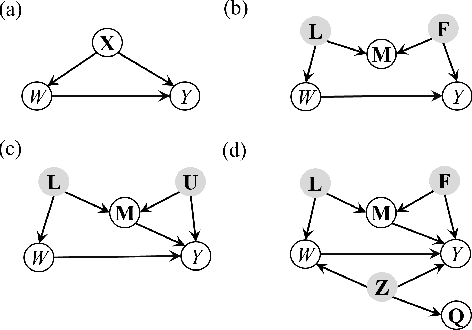
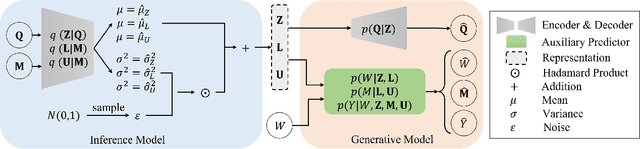
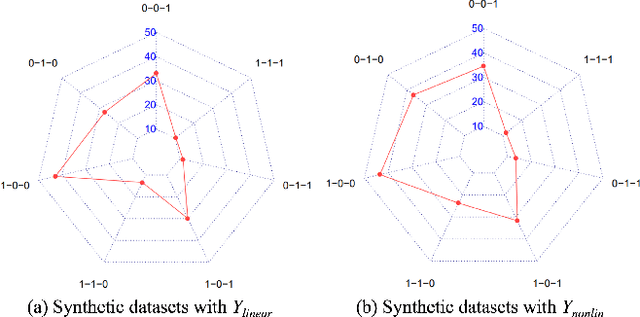
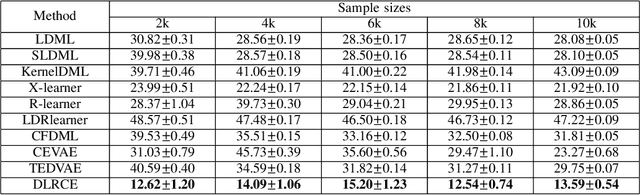
Abstract:In causal inference, it is a fundamental task to estimate the causal effect from observational data. However, latent confounders pose major challenges in causal inference in observational data, for example, confounding bias and M-bias. Recent data-driven causal effect estimators tackle the confounding bias problem via balanced representation learning, but assume no M-bias in the system, thus they fail to handle the M-bias. In this paper, we identify a challenging and unsolved problem caused by a variable that leads to confounding bias and M-bias simultaneously. To address this problem with co-occurring M-bias and confounding bias, we propose a novel Disentangled Latent Representation learning framework for learning latent representations from proxy variables for unbiased Causal effect Estimation (DLRCE) from observational data. Specifically, DLRCE learns three sets of latent representations from the measured proxy variables to adjust for the confounding bias and M-bias. Extensive experiments on both synthetic and three real-world datasets demonstrate that DLRCE significantly outperforms the state-of-the-art estimators in the case of the presence of both confounding bias and M-bias.
Research And Implementation Of Drug Target Interaction Confidence Measurement Method Based On Causal Intervention
May 31, 2023Abstract:The identification and discovery of drug-target Interaction (DTI) is an important step in the field of Drug research and development, which can help scientists discover new drugs and accelerate the development process. KnowledgeGraph and the related knowledge graph Embedding (KGE) model develop rapidly and show good performance in the field of drug discovery in recent years. In the task of drug target identification, the lack of authenticity and accuracy of the model will lead to the increase of misjudgment rate and the low efficiency of drug development. To solve the above problems, this study focused on the problem of drug target link prediction with knowledge mapping as the core technology, and adopted the confidence measurement method based on causal intervention to measure the triplet score, so as to improve the accuracy of drug target interaction prediction model. By comparing with the traditional Softmax and Sigmod confidence measurement methods on different KGE models, the results show that the confidence measurement method based on causal intervention can effectively improve the accuracy of DTI link prediction, especially for high-precision models. The predicted results are more conducive to guiding the design and development of followup experiments of drug development, so as to improve the efficiency of drug development.
hBert + BiasCorp -- Fighting Racism on the Web
Apr 06, 2021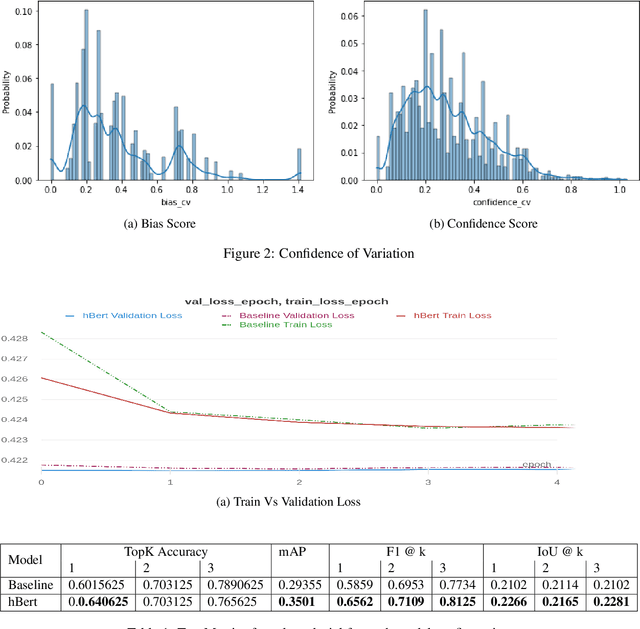
Abstract:Subtle and overt racism is still present both in physical and online communities today and has impacted many lives in different segments of the society. In this short piece of work, we present how we're tackling this societal issue with Natural Language Processing. We are releasing BiasCorp, a dataset containing 139,090 comments and news segment from three specific sources - Fox News, BreitbartNews and YouTube. The first batch (45,000 manually annotated) is ready for publication. We are currently in the final phase of manually labeling the remaining dataset using Amazon Mechanical Turk. BERT has been used widely in several downstream tasks. In this work, we present hBERT, where we modify certain layers of the pretrained BERT model with the new Hopfield Layer. hBert generalizes well across different distributions with the added advantage of a reduced model complexity. We are also releasing a JavaScript library and a Chrome Extension Application, to help developers make use of our trained model in web applications (say chat application) and for users to identify and report racially biased contents on the web respectively.
ConvPath: A Software Tool for Lung Adenocarcinoma Digital Pathological Image Analysis Aided by Convolutional Neural Network
Sep 20, 2018



Abstract:The spatial distributions of different types of cells could reveal a cancer cell growth pattern, its relationships with the tumor microenvironment and the immune response of the body, all of which represent key hallmarks of cancer. However, manually recognizing and localizing all the cells in pathology slides are almost impossible. In this study, we developed an automated cell type classification pipeline, ConvPath, which includes nuclei segmentation, convolutional neural network-based tumor, stromal and lymphocytes classification, and extraction of tumor microenvironment related features for lung cancer pathology images. The overall classification accuracy is 92.9% and 90.1% in training and independent testing datasets, respectively. By identifying cells and classifying cell types, this pipeline can convert a pathology image into a spatial map of tumor, stromal and lymphocyte cells. From this spatial map, we can extracted features that characterize the tumor micro-environment. Based on these features, we developed an image feature-based prognostic model and validated the model in two independent cohorts. The predicted risk group serves as an independent prognostic factor, after adjusting for clinical variables that include age, gender, smoking status, and stage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge