Bo Yao
Large language models enabled multiagent ensemble method for efficient EHR data labeling
Oct 21, 2024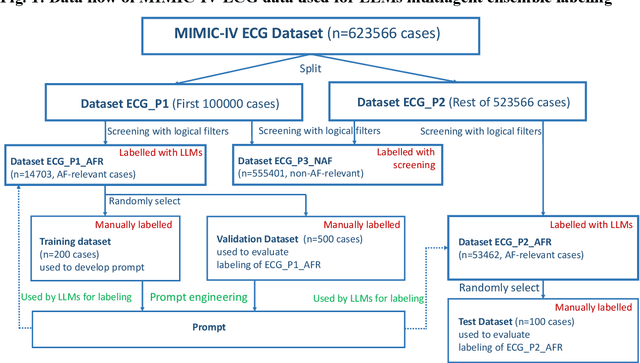
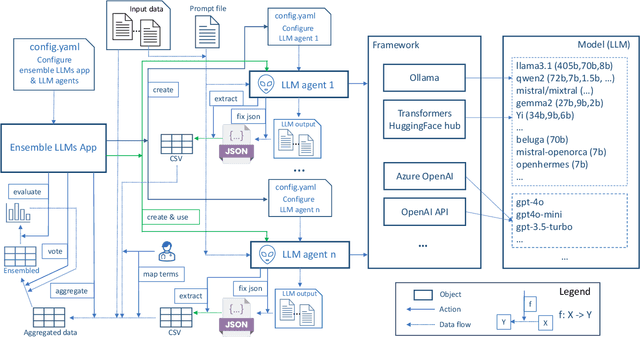
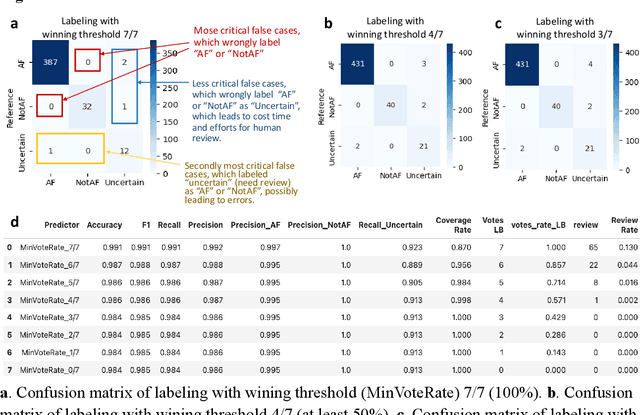
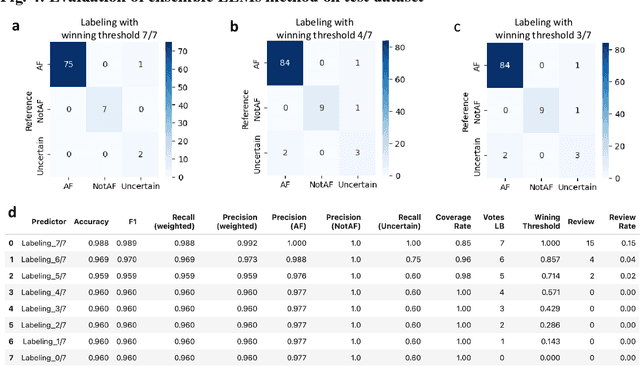
Abstract:This study introduces a novel multiagent ensemble method powered by LLMs to address a key challenge in ML - data labeling, particularly in large-scale EHR datasets. Manual labeling of such datasets requires domain expertise and is labor-intensive, time-consuming, expensive, and error-prone. To overcome this bottleneck, we developed an ensemble LLMs method and demonstrated its effectiveness in two real-world tasks: (1) labeling a large-scale unlabeled ECG dataset in MIMIC-IV; (2) identifying social determinants of health (SDOH) from the clinical notes of EHR. Trading off benefits and cost, we selected a pool of diverse open source LLMs with satisfactory performance. We treat each LLM's prediction as a vote and apply a mechanism of majority voting with minimal winning threshold for ensemble. We implemented an ensemble LLMs application for EHR data labeling tasks. By using the ensemble LLMs and natural language processing, we labeled MIMIC-IV ECG dataset of 623,566 ECG reports with an estimated accuracy of 98.2%. We applied the ensemble LLMs method to identify SDOH from social history sections of 1,405 EHR clinical notes, also achieving competitive performance. Our experiments show that the ensemble LLMs can outperform individual LLM even the best commercial one, and the method reduces hallucination errors. From the research, we found that (1) the ensemble LLMs method significantly reduces the time and effort required for labeling large-scale EHR data, automating the process with high accuracy and quality; (2) the method generalizes well to other text data labeling tasks, as shown by its application to SDOH identification; (3) the ensemble of a group of diverse LLMs can outperform or match the performance of the best individual LLM; and (4) the ensemble method substantially reduces hallucination errors. This approach provides a scalable and efficient solution to data-labeling challenges.
ConvPath: A Software Tool for Lung Adenocarcinoma Digital Pathological Image Analysis Aided by Convolutional Neural Network
Sep 20, 2018



Abstract:The spatial distributions of different types of cells could reveal a cancer cell growth pattern, its relationships with the tumor microenvironment and the immune response of the body, all of which represent key hallmarks of cancer. However, manually recognizing and localizing all the cells in pathology slides are almost impossible. In this study, we developed an automated cell type classification pipeline, ConvPath, which includes nuclei segmentation, convolutional neural network-based tumor, stromal and lymphocytes classification, and extraction of tumor microenvironment related features for lung cancer pathology images. The overall classification accuracy is 92.9% and 90.1% in training and independent testing datasets, respectively. By identifying cells and classifying cell types, this pipeline can convert a pathology image into a spatial map of tumor, stromal and lymphocyte cells. From this spatial map, we can extracted features that characterize the tumor micro-environment. Based on these features, we developed an image feature-based prognostic model and validated the model in two independent cohorts. The predicted risk group serves as an independent prognostic factor, after adjusting for clinical variables that include age, gender, smoking status, and stage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge