Xinliang Zhu
GENIUS: A Generative Framework for Universal Multimodal Search
Mar 25, 2025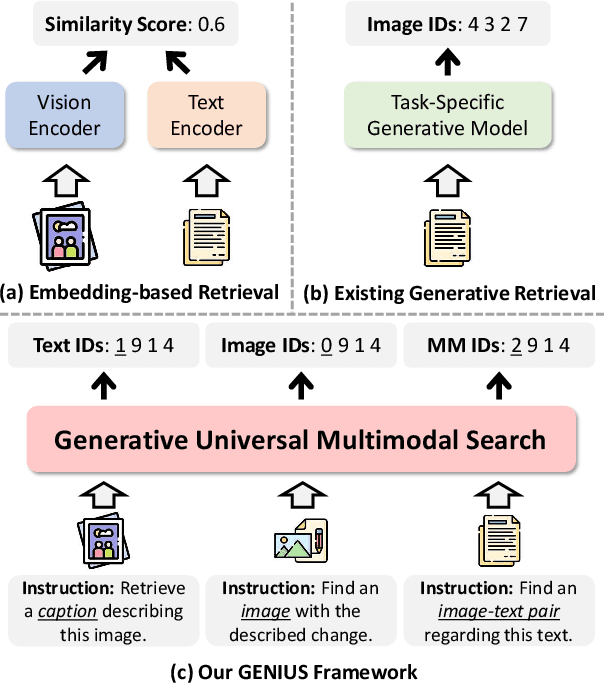
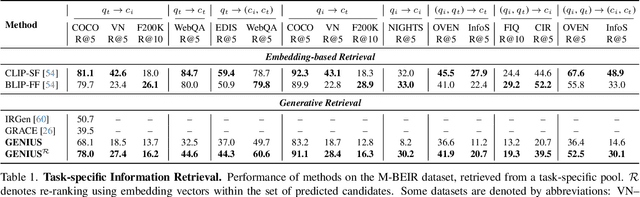
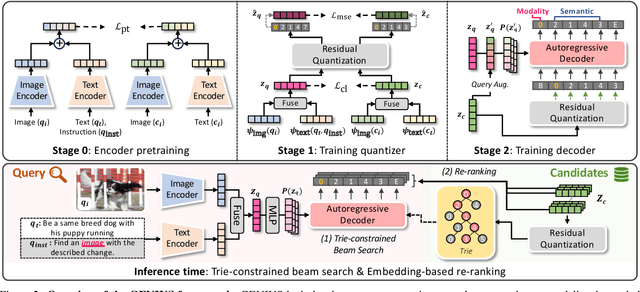
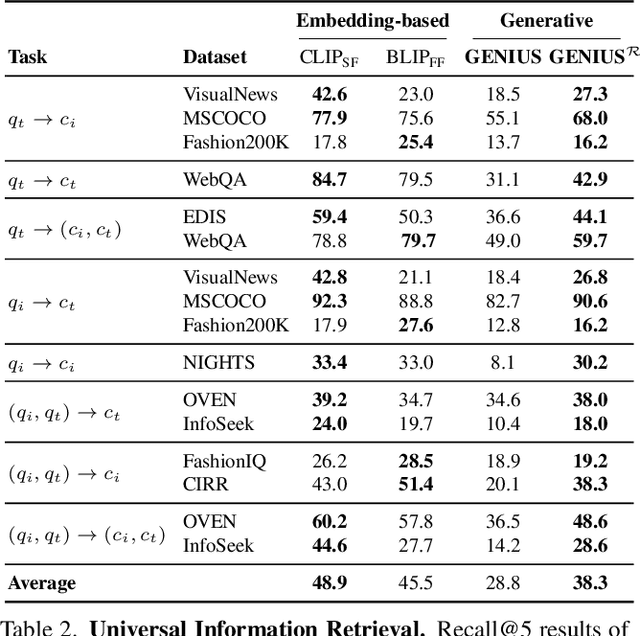
Abstract:Generative retrieval is an emerging approach in information retrieval that generates identifiers (IDs) of target data based on a query, providing an efficient alternative to traditional embedding-based retrieval methods. However, existing models are task-specific and fall short of embedding-based retrieval in performance. This paper proposes GENIUS, a universal generative retrieval framework supporting diverse tasks across multiple modalities and domains. At its core, GENIUS introduces modality-decoupled semantic quantization, transforming multimodal data into discrete IDs encoding both modality and semantics. Moreover, to enhance generalization, we propose a query augmentation that interpolates between a query and its target, allowing GENIUS to adapt to varied query forms. Evaluated on the M-BEIR benchmark, it surpasses prior generative methods by a clear margin. Unlike embedding-based retrieval, GENIUS consistently maintains high retrieval speed across database size, with competitive performance across multiple benchmarks. With additional re-ranking, GENIUS often achieves results close to those of embedding-based methods while preserving efficiency.
Bringing Multimodality to Amazon Visual Search System
Dec 17, 2024



Abstract:Image to image matching has been well studied in the computer vision community. Previous studies mainly focus on training a deep metric learning model matching visual patterns between the query image and gallery images. In this study, we show that pure image-to-image matching suffers from false positives caused by matching to local visual patterns. To alleviate this issue, we propose to leverage recent advances in vision-language pretraining research. Specifically, we introduce additional image-text alignment losses into deep metric learning, which serve as constraints to the image-to-image matching loss. With additional alignments between the text (e.g., product title) and image pairs, the model can learn concepts from both modalities explicitly, which avoids matching low-level visual features. We progressively develop two variants, a 3-tower and a 4-tower model, where the latter takes one more short text query input. Through extensive experiments, we show that this change leads to a substantial improvement to the image to image matching problem. We further leveraged this model for multimodal search, which takes both image and reformulation text queries to improve search quality. Both offline and online experiments show strong improvements on the main metrics. Specifically, we see 4.95% relative improvement on image matching click through rate with the 3-tower model and 1.13% further improvement from the 4-tower model.
Hierarchical Transformer for Survival Prediction Using Multimodality Whole Slide Images and Genomics
Nov 29, 2022Abstract:Learning good representation of giga-pixel level whole slide pathology images (WSI) for downstream tasks is critical. Previous studies employ multiple instance learning (MIL) to represent WSIs as bags of sampled patches because, for most occasions, only slide-level labels are available, and only a tiny region of the WSI is disease-positive area. However, WSI representation learning still remains an open problem due to: (1) patch sampling on a higher resolution may be incapable of depicting microenvironment information such as the relative position between the tumor cells and surrounding tissues, while patches at lower resolution lose the fine-grained detail; (2) extracting patches from giant WSI results in large bag size, which tremendously increases the computational cost. To solve the problems, this paper proposes a hierarchical-based multimodal transformer framework that learns a hierarchical mapping between pathology images and corresponding genes. Precisely, we randomly extract instant-level patch features from WSIs with different magnification. Then a co-attention mapping between imaging and genomics is learned to uncover the pairwise interaction and reduce the space complexity of imaging features. Such early fusion makes it computationally feasible to use MIL Transformer for the survival prediction task. Our architecture requires fewer GPU resources compared with benchmark methods while maintaining better WSI representation ability. We evaluate our approach on five cancer types from the Cancer Genome Atlas database and achieved an average c-index of $0.673$, outperforming the state-of-the-art multimodality methods.
Hierarchical Proxy-based Loss for Deep Metric Learning
Mar 25, 2021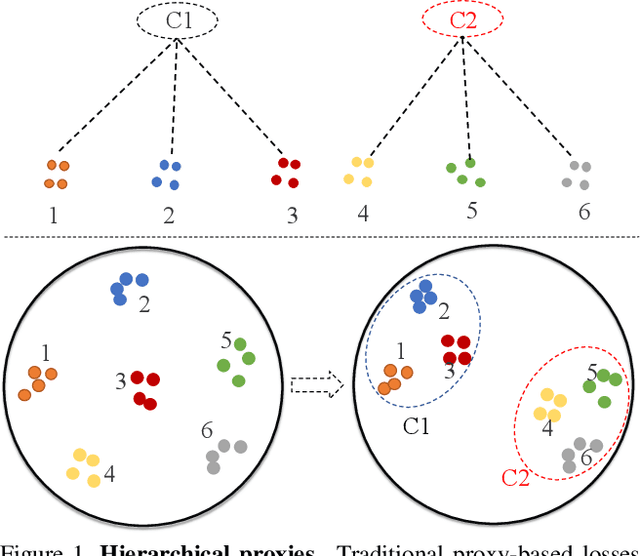

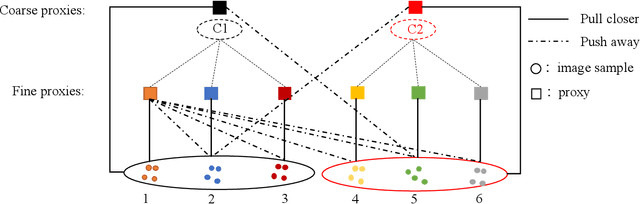

Abstract:Proxy-based metric learning losses are superior to pair-based losses due to their fast convergence and low training complexity. However, existing proxy-based losses focus on learning class-discriminative features while overlooking the commonalities shared across classes which are potentially useful in describing and matching samples. Moreover, they ignore the implicit hierarchy of categories in real-world datasets, where similar subordinate classes can be grouped together. In this paper, we present a framework that leverages this implicit hierarchy by imposing a hierarchical structure on the proxies and can be used with any existing proxy-based loss. This allows our model to capture both class-discriminative features and class-shared characteristics without breaking the implicit data hierarchy. We evaluate our method on five established image retrieval datasets such as In-Shop and SOP. Results demonstrate that our hierarchical proxy-based loss framework improves the performance of existing proxy-based losses, especially on large datasets which exhibit strong hierarchical structure.
Whole Slide Images based Cancer Survival Prediction using Attention Guided Deep Multiple Instance Learning Networks
Sep 23, 2020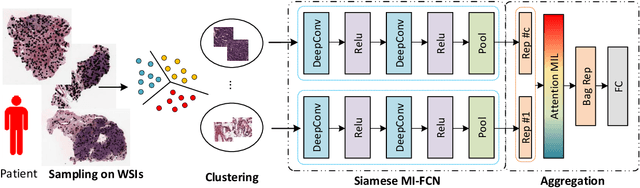
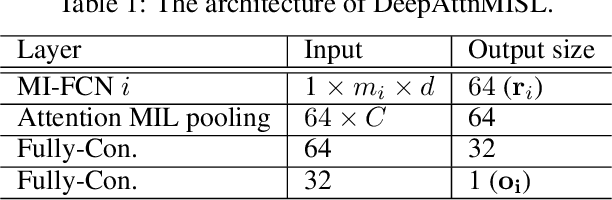
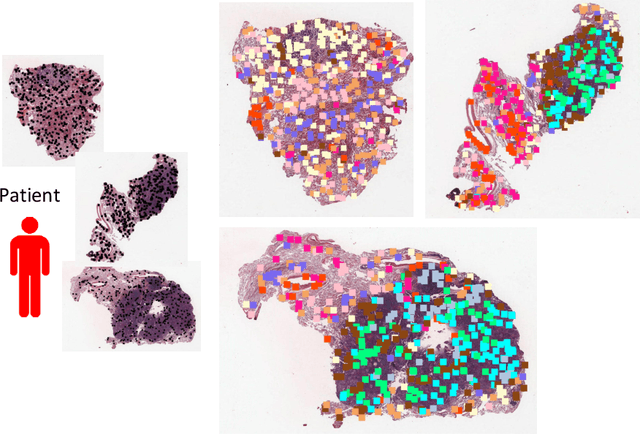
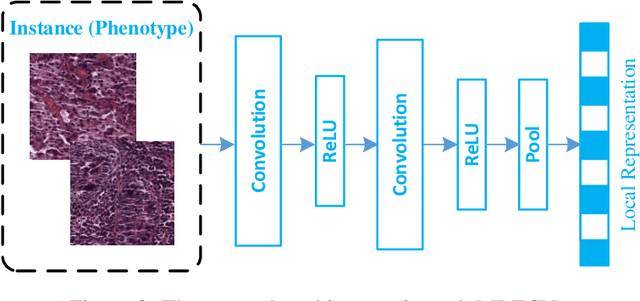
Abstract:Traditional image-based survival prediction models rely on discriminative patch labeling which make those methods not scalable to extend to large datasets. Recent studies have shown Multiple Instance Learning (MIL) framework is useful for histopathological images when no annotations are available in classification task. Different to the current image-based survival models that limit to key patches or clusters derived from Whole Slide Images (WSIs), we propose Deep Attention Multiple Instance Survival Learning (DeepAttnMISL) by introducing both siamese MI-FCN and attention-based MIL pooling to efficiently learn imaging features from the WSI and then aggregate WSI-level information to patient-level. Attention-based aggregation is more flexible and adaptive than aggregation techniques in recent survival models. We evaluated our methods on two large cancer whole slide images datasets and our results suggest that the proposed approach is more effective and suitable for large datasets and has better interpretability in locating important patterns and features that contribute to accurate cancer survival predictions. The proposed framework can also be used to assess individual patient's risk and thus assisting in delivering personalized medicine. Codes are available at https://github.com/uta-smile/DeepAttnMISL_MEDIA.
* 22 pages, 13 figures, published in Medical Image Analysis 65, 101789
Label-Driven Reconstruction for Domain Adaptation in Semantic Segmentation
Mar 10, 2020
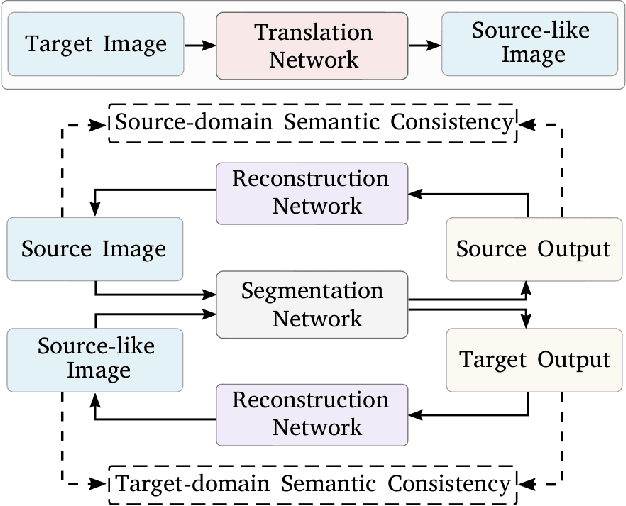
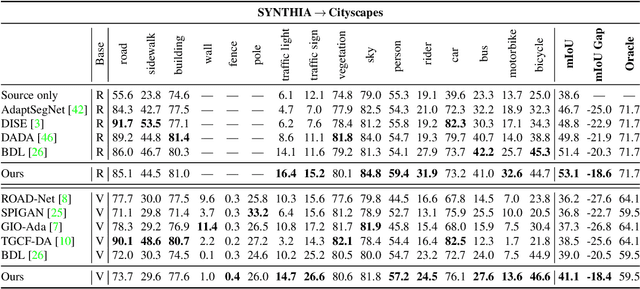
Abstract:Unsupervised domain adaptation enables to alleviate the need for pixel-wise annotation in the semantic segmentation. One of the most common strategies is to translate images from the source domain to the target domain and then align their marginal distributions in the feature space using adversarial learning. However, source-to-target translation enlarges the bias in translated images, owing to the dominant data size of the source domain. Furthermore, consistency of the joint distribution in source and target domains cannot be guaranteed through global feature alignment. Here, we present an innovative framework, designed to mitigate the image translation bias and align cross-domain features with the same category. This is achieved by 1) performing the target-to-source translation and 2) reconstructing both source and target images from their predicted labels. Extensive experiments on adapting from synthetic to real urban scene understanding demonstrate that our framework competes favorably against existing state-of-the-art methods.
Cohesion-based Online Actor-Critic Reinforcement Learning for mHealth Intervention
Aug 23, 2017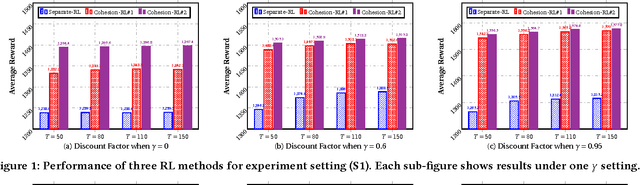

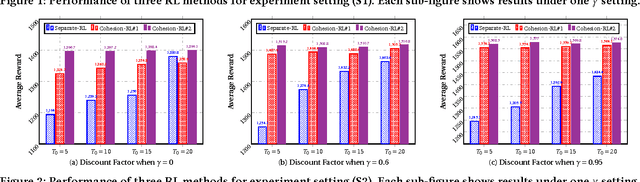
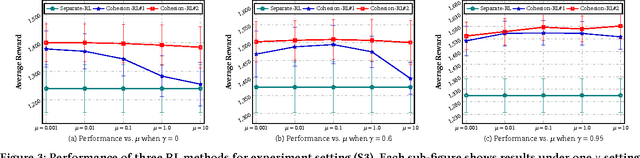
Abstract:In the wake of the vast population of smart device users worldwide, mobile health (mHealth) technologies are hopeful to generate positive and wide influence on people's health. They are able to provide flexible, affordable and portable health guides to device users. Current online decision-making methods for mHealth assume that the users are completely heterogeneous. They share no information among users and learn a separate policy for each user. However, data for each user is very limited in size to support the separate online learning, leading to unstable policies that contain lots of variances. Besides, we find the truth that a user may be similar with some, but not all, users, and connected users tend to have similar behaviors. In this paper, we propose a network cohesion constrained (actor-critic) Reinforcement Learning (RL) method for mHealth. The goal is to explore how to share information among similar users to better convert the limited user information into sharper learned policies. To the best of our knowledge, this is the first online actor-critic RL for mHealth and first network cohesion constrained (actor-critic) RL method in all applications. The network cohesion is important to derive effective policies. We come up with a novel method to learn the network by using the warm start trajectory, which directly reflects the users' property. The optimization of our model is difficult and very different from the general supervised learning due to the indirect observation of values. As a contribution, we propose two algorithms for the proposed online RLs. Apart from mHealth, the proposed methods can be easily applied or adapted to other health-related tasks. Extensive experiment results on the HeartSteps dataset demonstrates that in a variety of parameter settings, the proposed two methods obtain obvious improvements over the state-of-the-art methods.
Robust Contextual Bandit via the Capped-$\ell_{2}$ norm
Aug 17, 2017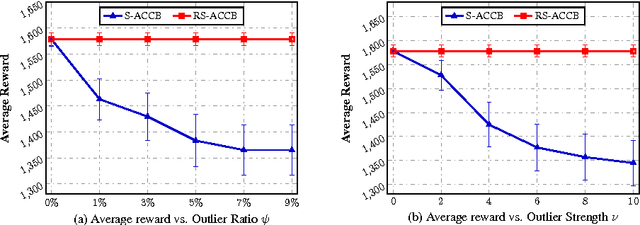
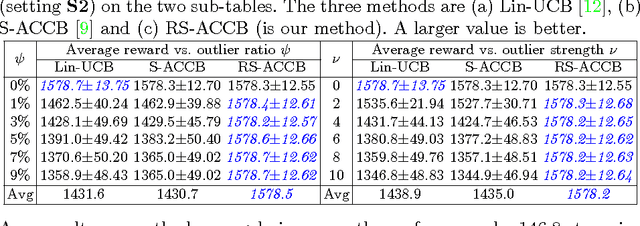
Abstract:This paper considers the actor-critic contextual bandit for the mobile health (mHealth) intervention. The state-of-the-art decision-making methods in mHealth generally assume that the noise in the dynamic system follows the Gaussian distribution. Those methods use the least-square-based algorithm to estimate the expected reward, which is prone to the existence of outliers. To deal with the issue of outliers, we propose a novel robust actor-critic contextual bandit method for the mHealth intervention. In the critic updating, the capped-$\ell_{2}$ norm is used to measure the approximation error, which prevents outliers from dominating our objective. A set of weights could be achieved from the critic updating. Considering them gives a weighted objective for the actor updating. It provides the badly noised sample in the critic updating with zero weights for the actor updating. As a result, the robustness of both actor-critic updating is enhanced. There is a key parameter in the capped-$\ell_{2}$ norm. We provide a reliable method to properly set it by making use of one of the most fundamental definitions of outliers in statistics. Extensive experiment results demonstrate that our method can achieve almost identical results compared with the state-of-the-art methods on the dataset without outliers and dramatically outperform them on the datasets noised by outliers.
10,000+ Times Accelerated Robust Subset Selection (ARSS)
Nov 17, 2014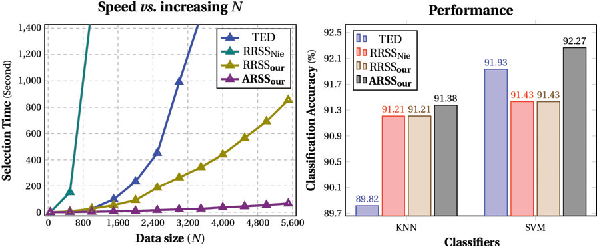
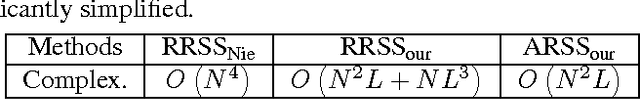
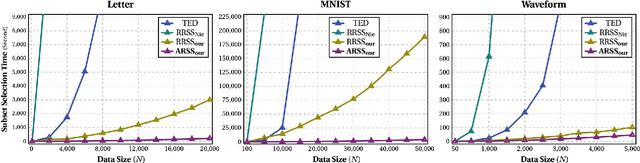

Abstract:Subset selection from massive data with noised information is increasingly popular for various applications. This problem is still highly challenging as current methods are generally slow in speed and sensitive to outliers. To address the above two issues, we propose an accelerated robust subset selection (ARSS) method. Specifically in the subset selection area, this is the first attempt to employ the $\ell_{p}(0<p\leq1)$-norm based measure for the representation loss, preventing large errors from dominating our objective. As a result, the robustness against outlier elements is greatly enhanced. Actually, data size is generally much larger than feature length, i.e. $N\gg L$. Based on this observation, we propose a speedup solver (via ALM and equivalent derivations) to highly reduce the computational cost, theoretically from $O(N^{4})$ to $O(N{}^{2}L)$. Extensive experiments on ten benchmark datasets verify that our method not only outperforms state of the art methods, but also runs 10,000+ times faster than the most related method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge