Xavier Intes
JADAI: Jointly Amortizing Adaptive Design and Bayesian Inference
Dec 28, 2025Abstract:We consider problems of parameter estimation where design variables can be actively optimized to maximize information gain. To this end, we introduce JADAI, a framework that jointly amortizes Bayesian adaptive design and inference by training a policy, a history network, and an inference network end-to-end. The networks minimize a generic loss that aggregates incremental reductions in posterior error along experimental sequences. Inference networks are instantiated with diffusion-based posterior estimators that can approximate high-dimensional and multimodal posteriors at every experimental step. Across standard adaptive design benchmarks, JADAI achieves superior or competitive performance.
Enhancing Fluorescence Lifetime Parameter Estimation Accuracy with Differential Transformer Based Deep Learning Model Incorporating Pixelwise Instrument Response Function
Nov 25, 2024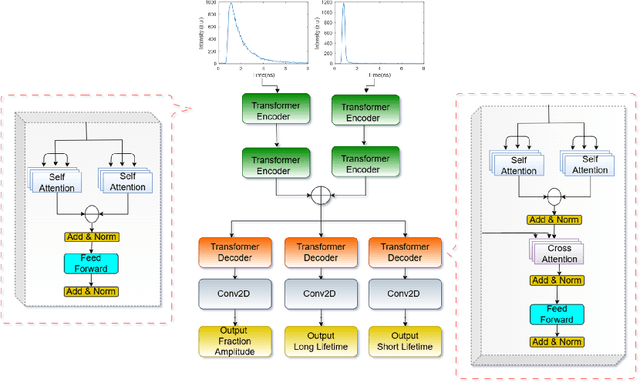
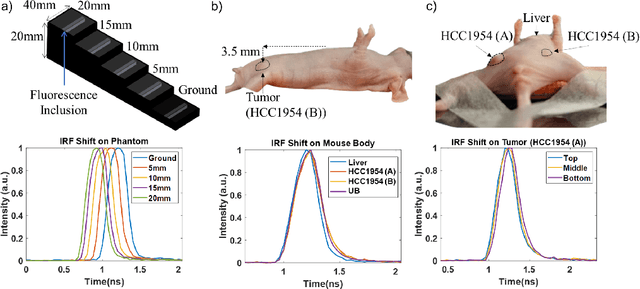
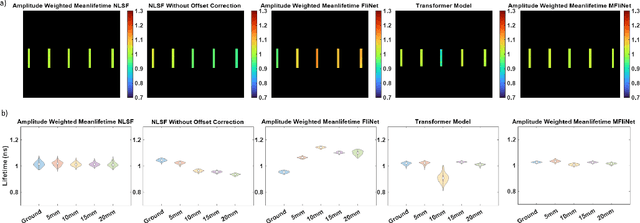
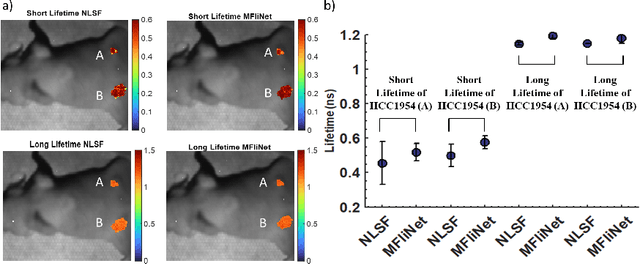
Abstract:Fluorescence lifetime imaging (FLI) is an important molecular imaging modality that can provide unique information for biomedical applications. FLI is based on acquiring and processing photon time of arrival histograms. The shape and temporal offset of these histograms depends on many factors, such as the instrument response function (IRF), optical properties, and the topographic profile of the sample. Several inverse solver analytical methods have been developed to compute the underlying fluorescence lifetime parameters, but most of them are computationally expensive and time-consuming. Thus, deep learning (DL) algorithms have progressively replaced computation methods in fluorescence lifetime parameter estimation. Often, DL models are trained with simple datasets either generated through simulation or a simple experiment where the fluorophore surface profile is mostly flat; therefore, DL models often do not perform well on samples with complex surface profiles such as ex-vivo organs or in-vivo whole intact animals. Herein, we introduce a new DL architecture using state-of-the-art Differential Transformer encoder-decoder architecture, MFliNet (Macroscopic FLI Network), that takes an additional input of IRF together with TPSF, addressing discrepancies in the photon time-of-arrival distribution. We demonstrate the model's performance through carefully designed, complex tissue-mimicking phantoms and preclinical in-vivo cancer xenograft experiments.
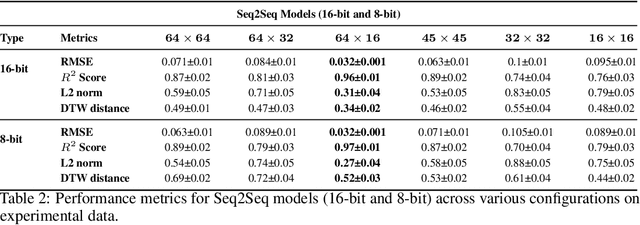
Unlocking Real-Time Fluorescence Lifetime Imaging: Multi-Pixel Parallelism for FPGA-Accelerated Processing
Oct 09, 2024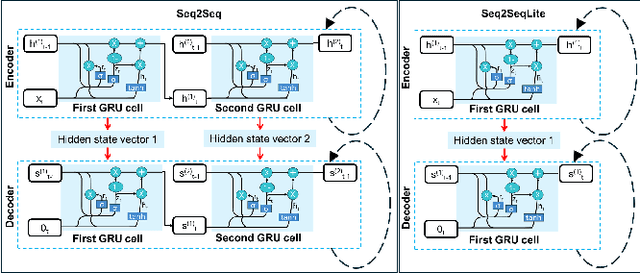
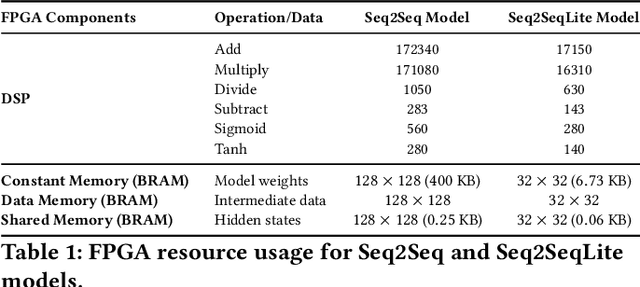

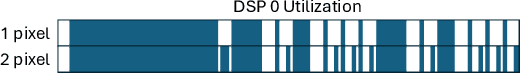
Abstract:Fluorescence lifetime imaging (FLI) is a widely used technique in the biomedical field for measuring the decay times of fluorescent molecules, providing insights into metabolic states, protein interactions, and ligand-receptor bindings. However, its broader application in fast biological processes, such as dynamic activity monitoring, and clinical use, such as in guided surgery, is limited by long data acquisition times and computationally demanding data processing. While deep learning has reduced post-processing times, time-resolved data acquisition remains a bottleneck for real-time applications. To address this, we propose a method to achieve real-time FLI using an FPGA-based hardware accelerator. Specifically, we implemented a GRU-based sequence-to-sequence (Seq2Seq) model on an FPGA board compatible with time-resolved cameras. The GRU model balances accurate processing with the resource constraints of FPGAs, which have limited DSP units and BRAM. The limited memory and computational resources on the FPGA require efficient scheduling of operations and memory allocation to deploy deep learning models for low-latency applications. We address these challenges by using STOMP, a queue-based discrete-event simulator that automates and optimizes task scheduling and memory management on hardware. By integrating a GRU-based Seq2Seq model and its compressed version, called Seq2SeqLite, generated through knowledge distillation, we were able to process multiple pixels in parallel, reducing latency compared to sequential processing. We explore various levels of parallelism to achieve an optimal balance between performance and resource utilization. Our results indicate that the proposed techniques achieved a 17.7x and 52.0x speedup over manual scheduling for the Seq2Seq model and the Seq2SeqLite model, respectively.
Compressing Recurrent Neural Networks for FPGA-accelerated Implementation in Fluorescence Lifetime Imaging
Oct 01, 2024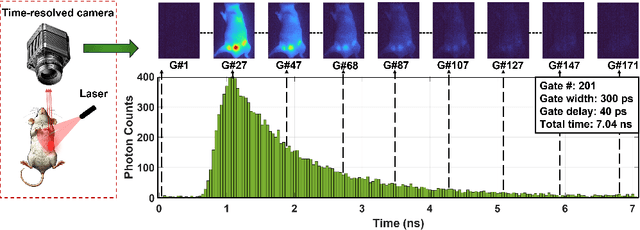

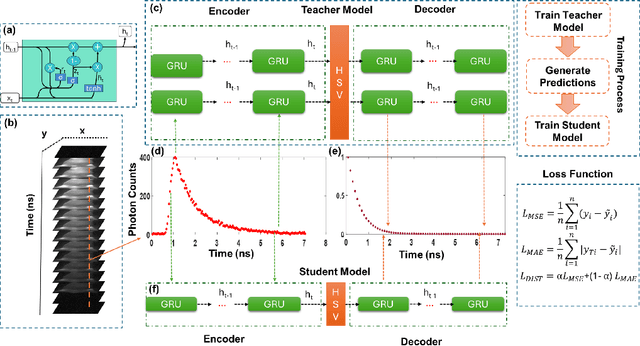
Abstract:Fluorescence lifetime imaging (FLI) is an important technique for studying cellular environments and molecular interactions, but its real-time application is limited by slow data acquisition, which requires capturing large time-resolved images and complex post-processing using iterative fitting algorithms. Deep learning (DL) models enable real-time inference, but can be computationally demanding due to complex architectures and large matrix operations. This makes DL models ill-suited for direct implementation on field-programmable gate array (FPGA)-based camera hardware. Model compression is thus crucial for practical deployment for real-time inference generation. In this work, we focus on compressing recurrent neural networks (RNNs), which are well-suited for FLI time-series data processing, to enable deployment on resource-constrained FPGA boards. We perform an empirical evaluation of various compression techniques, including weight reduction, knowledge distillation (KD), post-training quantization (PTQ), and quantization-aware training (QAT), to reduce model size and computational load while preserving inference accuracy. Our compressed RNN model, Seq2SeqLite, achieves a balance between computational efficiency and prediction accuracy, particularly at 8-bit precision. By applying KD, the model parameter size was reduced by 98\% while retaining performance, making it suitable for concurrent real-time FLI analysis on FPGA during data capture. This work represents a big step towards integrating hardware-accelerated real-time FLI analysis for fast biological processes.
Cognitive-Motor Integration in Assessing Bimanual Motor Skills
Apr 16, 2024



Abstract:Accurate assessment of bimanual motor skills is essential across various professions, yet, traditional methods often rely on subjective assessments or focus solely on motor actions, overlooking the integral role of cognitive processes. This study introduces a novel approach by leveraging deep neural networks (DNNs) to analyze and integrate both cognitive decision-making and motor execution. We tested this methodology by assessing laparoscopic surgery skills within the Fundamentals of Laparoscopic Surgery program, which is a prerequisite for general surgery certification. Utilizing video capture of motor actions and non-invasive functional near-infrared spectroscopy (fNIRS) for measuring neural activations, our approach precisely classifies subjects by expertise level and predicts FLS behavioral performance scores, significantly surpassing traditional single-modality assessments.
Roadmap on Deep Learning for Microscopy
Mar 07, 2023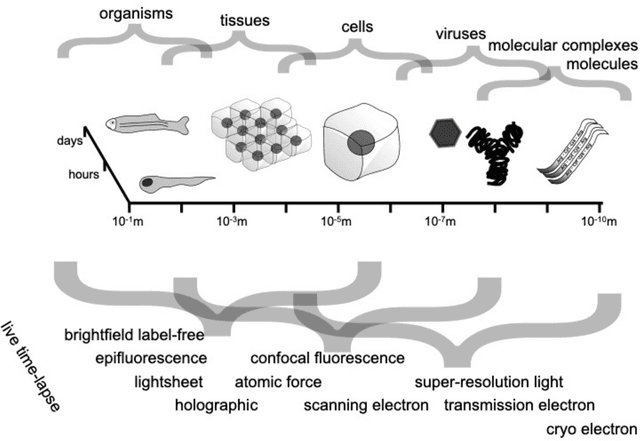
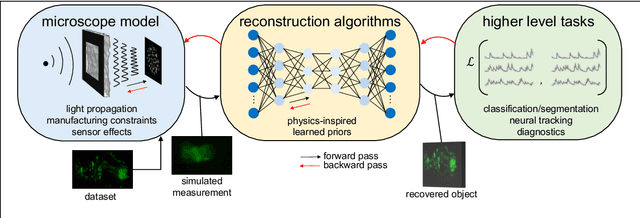
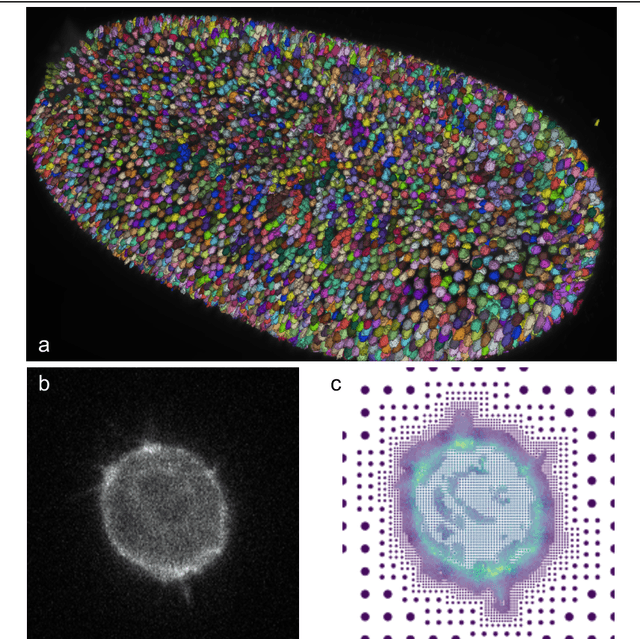
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
One-shot domain adaptation in video-based assessment of surgical skills
Dec 16, 2022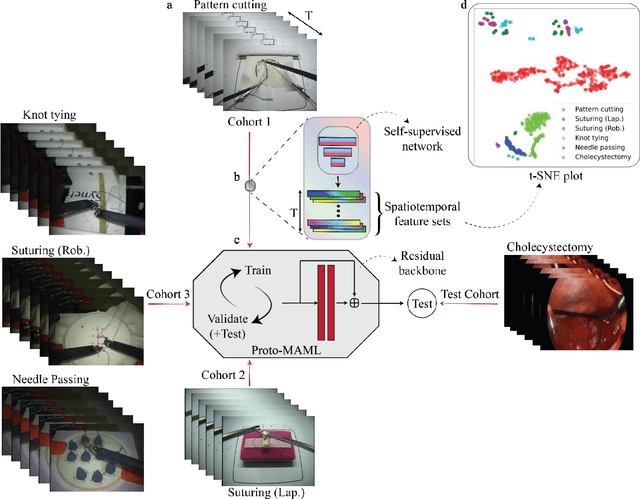
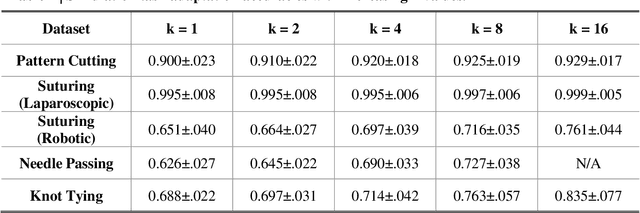
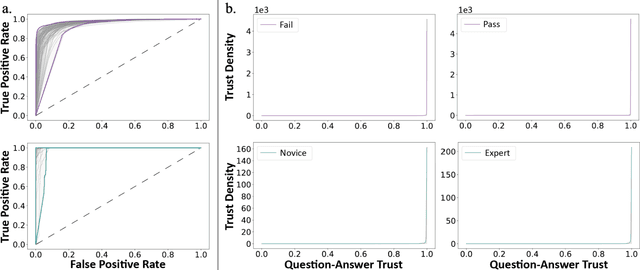
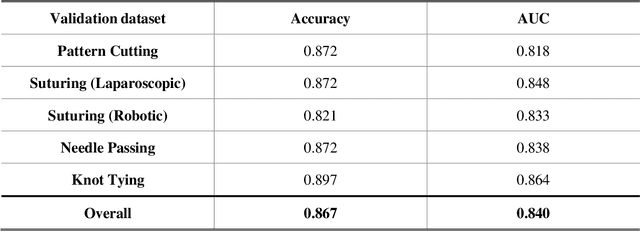
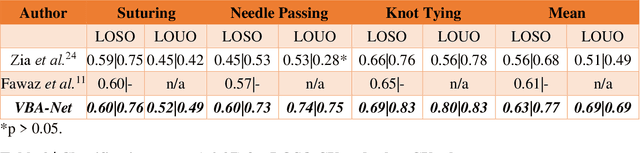
Abstract:Deep Learning (DL) has achieved automatic and objective assessment of surgical skills. However, DL models are data-hungry and restricted to their training domain. This prevents them from transitioning to new tasks where data is limited. Hence, domain adaptation is crucial to implement DL in real life. Here, we propose a meta-learning model, A-VBANet, that can deliver domain-agnostic surgical skill classification via one-shot learning. We develop the A-VBANet on five laparoscopic and robotic surgical simulators. Additionally, we test it on operating room (OR) videos of laparoscopic cholecystectomy. Our model successfully adapts with accuracies up to 99.5% in one-shot and 99.9% in few-shot settings for simulated tasks and 89.7% for laparoscopic cholecystectomy. For the first time, we provide a domain-agnostic procedure for video-based assessment of surgical skills. A significant implication of this approach is that it allows the use of data from surgical simulators to assess performance in the operating room.
Video-based Formative and Summative Assessment of Surgical Tasks using Deep Learning
Mar 17, 2022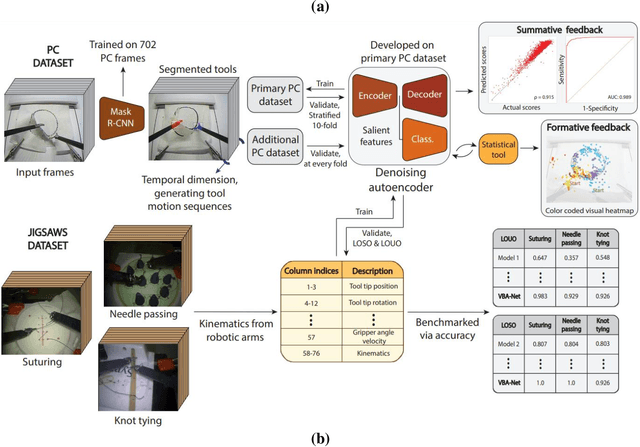
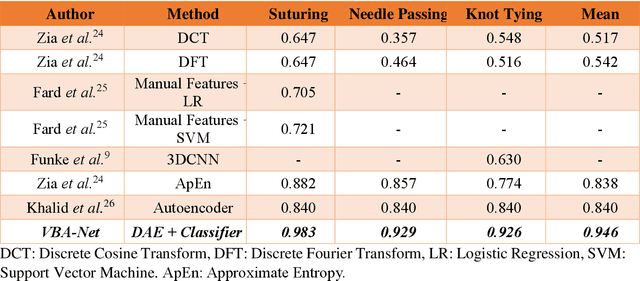
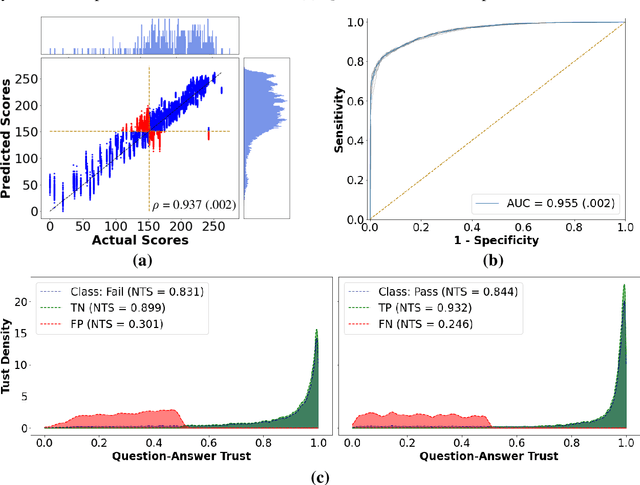
Abstract:To ensure satisfactory clinical outcomes, surgical skill assessment must be objective, time-efficient, and preferentially automated - none of which is currently achievable. Video-based assessment (VBA) is being deployed in intraoperative and simulation settings to evaluate technical skill execution. However, VBA remains manually- and time-intensive and prone to subjective interpretation and poor inter-rater reliability. Herein, we propose a deep learning (DL) model that can automatically and objectively provide a high-stakes summative assessment of surgical skill execution based on video feeds and low-stakes formative assessment to guide surgical skill acquisition. Formative assessment is generated using heatmaps of visual features that correlate with surgical performance. Hence, the DL model paves the way to the quantitative and reproducible evaluation of surgical tasks from videos with the potential for broad dissemination in surgical training, certification, and credentialing.
Deep Neural Networks for the Assessment of Surgical Skills: A Systematic Review
Mar 03, 2021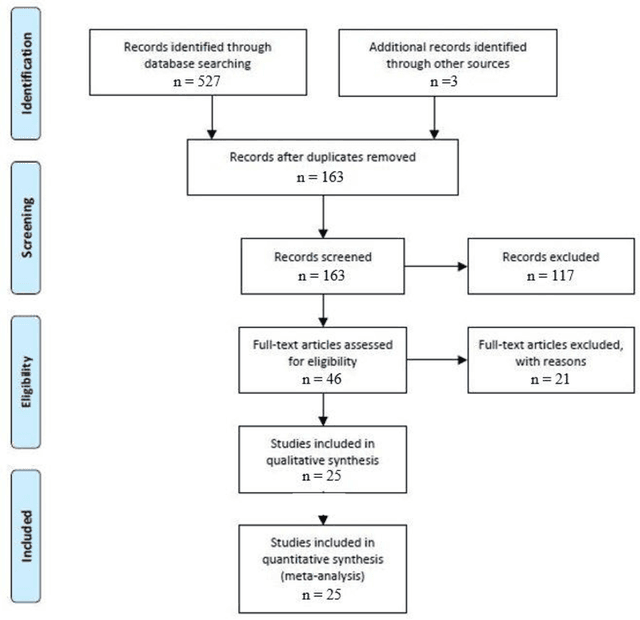
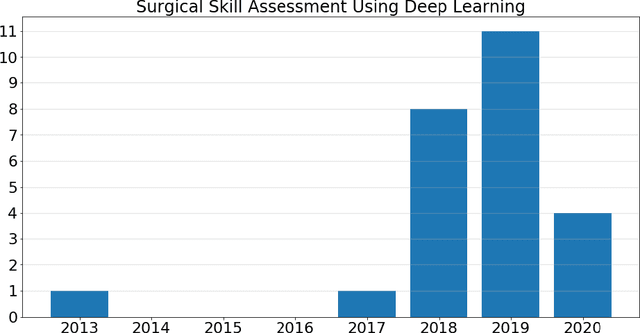
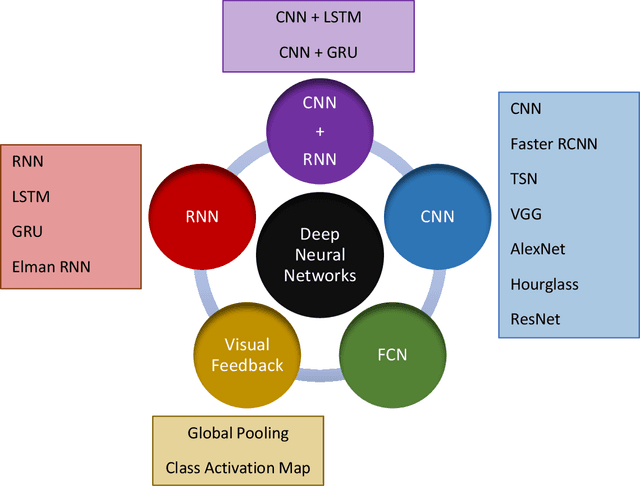
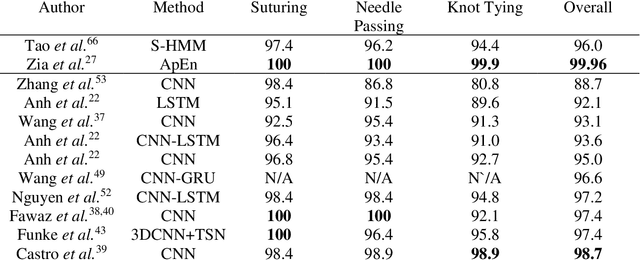
Abstract:Surgical training in medical school residency programs has followed the apprenticeship model. The learning and assessment process is inherently subjective and time-consuming. Thus, there is a need for objective methods to assess surgical skills. Here, we use the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines to systematically survey the literature on the use of Deep Neural Networks for automated and objective surgical skill assessment, with a focus on kinematic data as putative markers of surgical competency. There is considerable recent interest in deep neural networks (DNN) due to the availability of powerful algorithms, multiple datasets, some of which are publicly available, as well as efficient computational hardware to train and host them. We have reviewed 530 papers, of which we selected 25 for this systematic review. Based on this review, we concluded that DNNs are powerful tools for automated, objective surgical skill assessment using both kinematic and video data. The field would benefit from large, publicly available, annotated datasets that are representative of the surgical trainee and expert demographics and multimodal data beyond kinematics and videos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge