Wei Lou
Cell Graph Transformer for Nuclei Classification
Feb 20, 2024Abstract:Nuclei classification is a critical step in computer-aided diagnosis with histopathology images. In the past, various methods have employed graph neural networks (GNN) to analyze cell graphs that model inter-cell relationships by considering nuclei as vertices. However, they are limited by the GNN mechanism that only passes messages among local nodes via fixed edges. To address the issue, we develop a cell graph transformer (CGT) that treats nodes and edges as input tokens to enable learnable adjacency and information exchange among all nodes. Nevertheless, training the transformer with a cell graph presents another challenge. Poorly initialized features can lead to noisy self-attention scores and inferior convergence, particularly when processing the cell graphs with numerous connections. Thus, we further propose a novel topology-aware pretraining method that leverages a graph convolutional network (GCN) to learn a feature extractor. The pre-trained features may suppress unreasonable correlations and hence ease the finetuning of CGT. Experimental results suggest that the proposed cell graph transformer with topology-aware pretraining significantly improves the nuclei classification results, and achieves the state-of-the-art performance. Code and models are available at https://github.com/lhaof/CGT
Diffusion-based Data Augmentation for Nuclei Image Segmentation
Oct 22, 2023Abstract:Nuclei segmentation is a fundamental but challenging task in the quantitative analysis of histopathology images. Although fully-supervised deep learning-based methods have made significant progress, a large number of labeled images are required to achieve great segmentation performance. Considering that manually labeling all nuclei instances for a dataset is inefficient, obtaining a large-scale human-annotated dataset is time-consuming and labor-intensive. Therefore, augmenting a dataset with only a few labeled images to improve the segmentation performance is of significant research and application value. In this paper, we introduce the first diffusion-based augmentation method for nuclei segmentation. The idea is to synthesize a large number of labeled images to facilitate training the segmentation model. To achieve this, we propose a two-step strategy. In the first step, we train an unconditional diffusion model to synthesize the Nuclei Structure that is defined as the representation of pixel-level semantic and distance transform. Each synthetic nuclei structure will serve as a constraint on histopathology image synthesis and is further post-processed to be an instance map. In the second step, we train a conditioned diffusion model to synthesize histopathology images based on nuclei structures. The synthetic histopathology images paired with synthetic instance maps will be added to the real dataset for training the segmentation model. The experimental results show that by augmenting 10% labeled real dataset with synthetic samples, one can achieve comparable segmentation results with the fully-supervised baseline.
Multi-stream Cell Segmentation with Low-level Cues for Multi-modality Images
Oct 22, 2023Abstract:Cell segmentation for multi-modal microscopy images remains a challenge due to the complex textures, patterns, and cell shapes in these images. To tackle the problem, we first develop an automatic cell classification pipeline to label the microscopy images based on their low-level image characteristics, and then train a classification model based on the category labels. Afterward, we train a separate segmentation model for each category using the images in the corresponding category. Besides, we further deploy two types of segmentation models to segment cells with roundish and irregular shapes respectively. Moreover, an efficient and powerful backbone model is utilized to enhance the efficiency of our segmentation model. Evaluated on the Tuning Set of NeurIPS 2022 Cell Segmentation Challenge, our method achieves an F1-score of 0.8795 and the running time for all cases is within the time tolerance.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
Structure Embedded Nucleus Classification for Histopathology Images
Feb 22, 2023Abstract:Nuclei classification provides valuable information for histopathology image analysis. However, the large variations in the appearance of different nuclei types cause difficulties in identifying nuclei. Most neural network based methods are affected by the local receptive field of convolutions, and pay less attention to the spatial distribution of nuclei or the irregular contour shape of a nucleus. In this paper, we first propose a novel polygon-structure feature learning mechanism that transforms a nucleus contour into a sequence of points sampled in order, and employ a recurrent neural network that aggregates the sequential change in distance between key points to obtain learnable shape features. Next, we convert a histopathology image into a graph structure with nuclei as nodes, and build a graph neural network to embed the spatial distribution of nuclei into their representations. To capture the correlations between the categories of nuclei and their surrounding tissue patterns, we further introduce edge features that are defined as the background textures between adjacent nuclei. Lastly, we integrate both polygon and graph structure learning mechanisms into a whole framework that can extract intra and inter-nucleus structural characteristics for nuclei classification. Experimental results show that the proposed framework achieves significant improvements compared to the state-of-the-art methods.
Which Pixel to Annotate: a Label-Efficient Nuclei Segmentation Framework
Dec 20, 2022



Abstract:Recently deep neural networks, which require a large amount of annotated samples, have been widely applied in nuclei instance segmentation of H\&E stained pathology images. However, it is inefficient and unnecessary to label all pixels for a dataset of nuclei images which usually contain similar and redundant patterns. Although unsupervised and semi-supervised learning methods have been studied for nuclei segmentation, very few works have delved into the selective labeling of samples to reduce the workload of annotation. Thus, in this paper, we propose a novel full nuclei segmentation framework that chooses only a few image patches to be annotated, augments the training set from the selected samples, and achieves nuclei segmentation in a semi-supervised manner. In the proposed framework, we first develop a novel consistency-based patch selection method to determine which image patches are the most beneficial to the training. Then we introduce a conditional single-image GAN with a component-wise discriminator, to synthesize more training samples. Lastly, our proposed framework trains an existing segmentation model with the above augmented samples. The experimental results show that our proposed method could obtain the same-level performance as a fully-supervised baseline by annotating less than 5% pixels on some benchmarks.
Fast Heterogeneous Federated Learning with Hybrid Client Selection
Aug 16, 2022



Abstract:Client selection schemes are widely adopted to handle the communication-efficient problems in recent studies of Federated Learning (FL). However, the large variance of the model updates aggregated from the randomly-selected unrepresentative subsets directly slows the FL convergence. We present a novel clustering-based client selection scheme to accelerate the FL convergence by variance reduction. Simple yet effective schemes are designed to improve the clustering effect and control the effect fluctuation, therefore, generating the client subset with certain representativeness of sampling. Theoretically, we demonstrate the improvement of the proposed scheme in variance reduction. We also present the tighter convergence guarantee of the proposed method thanks to the variance reduction. Experimental results confirm the exceed efficiency of our scheme compared to alternatives.
Variance-Reduced Heterogeneous Federated Learning via Stratified Client Selection
Jan 15, 2022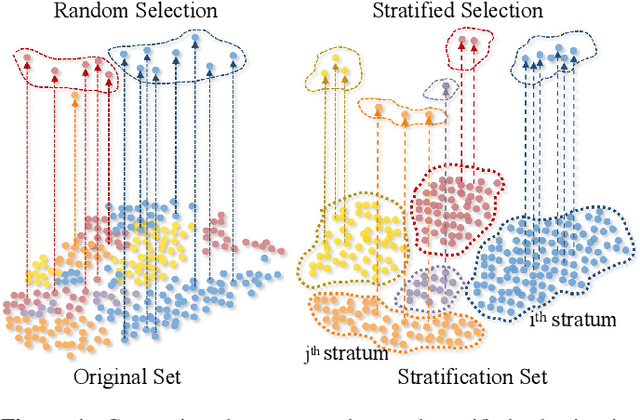
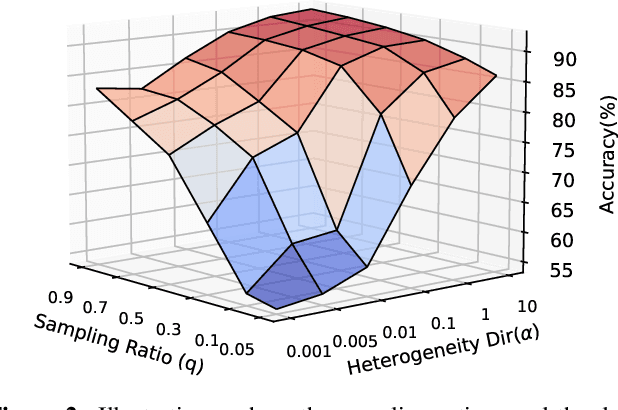
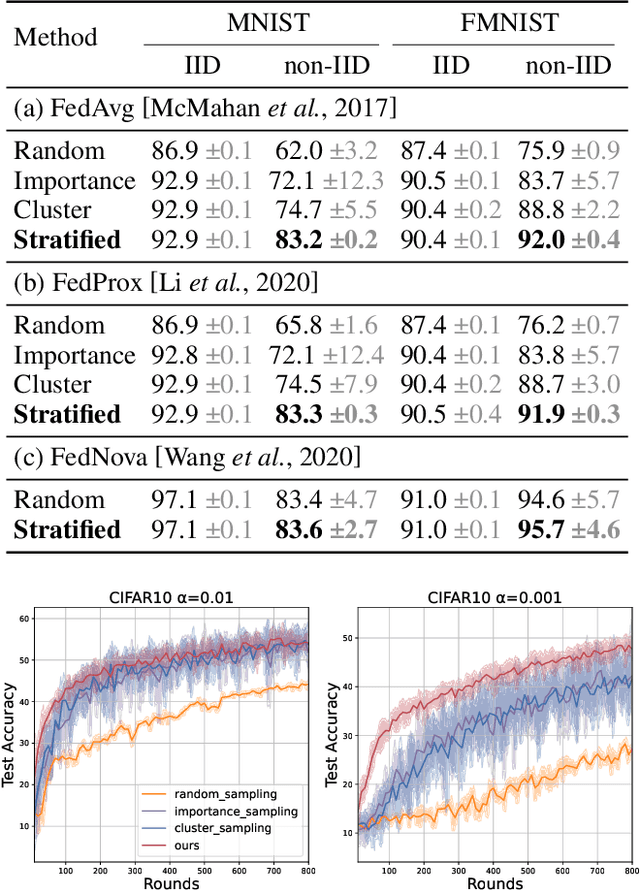
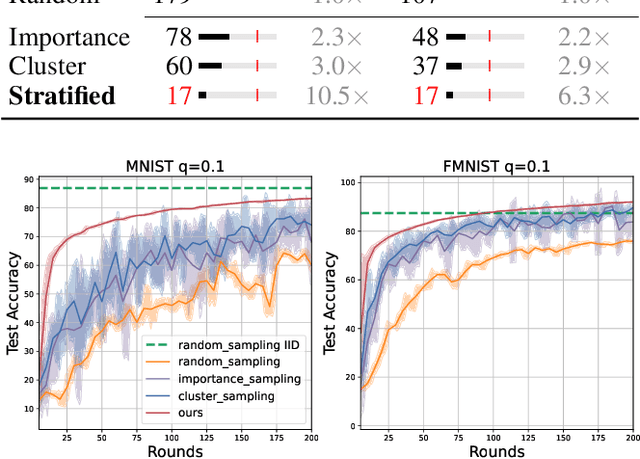
Abstract:Client selection strategies are widely adopted to handle the communication-efficient problem in recent studies of Federated Learning (FL). However, due to the large variance of the selected subset's update, prior selection approaches with a limited sampling ratio cannot perform well on convergence and accuracy in heterogeneous FL. To address this problem, in this paper, we propose a novel stratified client selection scheme to reduce the variance for the pursuit of better convergence and higher accuracy. Specifically, to mitigate the impact of heterogeneity, we develop stratification based on clients' local data distribution to derive approximate homogeneous strata for better selection in each stratum. Concentrating on a limited sampling ratio scenario, we next present an optimized sample size allocation scheme by considering the diversity of stratum's variability, with the promise of further variance reduction. Theoretically, we elaborate the explicit relation among different selection schemes with regard to variance, under heterogeneous settings, we demonstrate the effectiveness of our selection scheme. Experimental results confirm that our approach not only allows for better performance relative to state-of-the-art methods but also is compatible with prevalent FL algorithms.
Dynamic-OFA: Runtime DNN Architecture Switching for Performance Scaling on Heterogeneous Embedded Platforms
May 11, 2021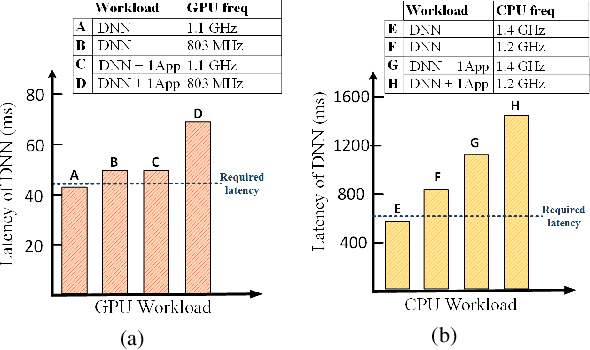

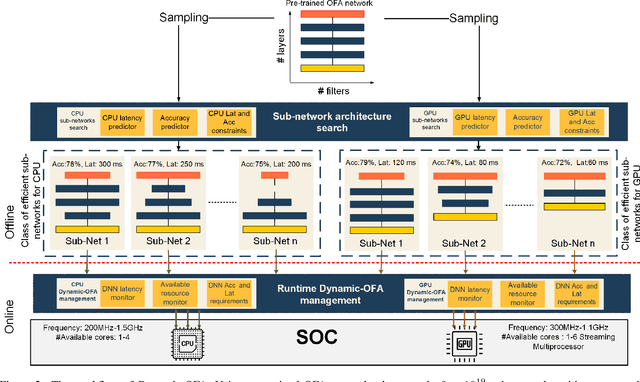
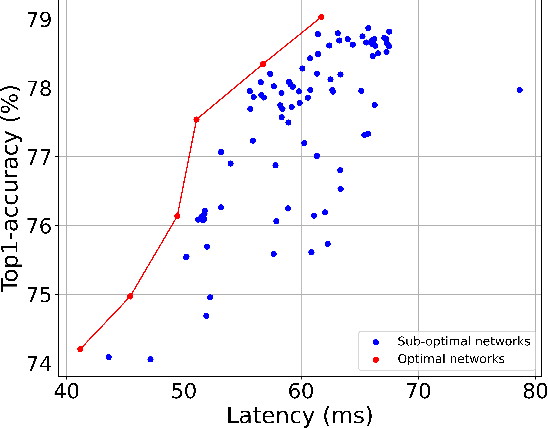
Abstract:Mobile and embedded platforms are increasingly required to efficiently execute computationally demanding DNNs across heterogeneous processing elements. At runtime, the available hardware resources to DNNs can vary considerably due to other concurrently running applications. The performance requirements of the applications could also change under different scenarios. To achieve the desired performance, dynamic DNNs have been proposed in which the number of channels/layers can be scaled in real time to meet different requirements under varying resource constraints. However, the training process of such dynamic DNNs can be costly, since platform-aware models of different deployment scenarios must be retrained to become dynamic. This paper proposes Dynamic-OFA, a novel dynamic DNN approach for state-of-the-art platform-aware NAS models (i.e. Once-for-all network (OFA)). Dynamic-OFA pre-samples a family of sub-networks from a static OFA backbone model, and contains a runtime manager to choose different sub-networks under different runtime environments. As such, Dynamic-OFA does not need the traditional dynamic DNN training pipeline. Compared to the state-of-the-art, our experimental results using ImageNet on a Jetson Xavier NX show that the approach is up to 3.5x (CPU), 2.4x (GPU) faster for similar ImageNet Top-1 accuracy, or 3.8% (CPU), 5.1% (GPU) higher accuracy at similar latency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge