Haofeng Li
JieHua Paintings Style Feature Extracting Model using Stable Diffusion with ControlNet
Aug 21, 2024


Abstract:This study proposes a novel approach to extract stylistic features of Jiehua: the utilization of the Fine-tuned Stable Diffusion Model with ControlNet (FSDMC) to refine depiction techniques from artists' Jiehua. The training data for FSDMC is based on the opensource Jiehua artist's work collected from the Internet, which were subsequently manually constructed in the format of (Original Image, Canny Edge Features, Text Prompt). By employing the optimal hyperparameters identified in this paper, it was observed FSDMC outperforms CycleGAN, another mainstream style transfer model. FSDMC achieves FID of 3.27 on the dataset and also surpasses CycleGAN in terms of expert evaluation. This not only demonstrates the model's high effectiveness in extracting Jiehua's style features, but also preserves the original pre-trained semantic information. The findings of this study suggest that the application of FSDMC with appropriate hyperparameters can enhance the efficacy of the Stable Diffusion Model in the field of traditional art style migration tasks, particularly within the context of Jiehua.
Diffuse-UDA: Addressing Unsupervised Domain Adaptation in Medical Image Segmentation with Appearance and Structure Aligned Diffusion Models
Aug 12, 2024Abstract:The scarcity and complexity of voxel-level annotations in 3D medical imaging present significant challenges, particularly due to the domain gap between labeled datasets from well-resourced centers and unlabeled datasets from less-resourced centers. This disparity affects the fairness of artificial intelligence algorithms in healthcare. We introduce Diffuse-UDA, a novel method leveraging diffusion models to tackle Unsupervised Domain Adaptation (UDA) in medical image segmentation. Diffuse-UDA generates high-quality image-mask pairs with target domain characteristics and various structures, thereby enhancing UDA tasks. Initially, pseudo labels for target domain samples are generated. Subsequently, a specially tailored diffusion model, incorporating deformable augmentations, is trained on image-label or image-pseudo-label pairs from both domains. Finally, source domain labels guide the diffusion model to generate image-label pairs for the target domain. Comprehensive evaluations on several benchmarks demonstrate that Diffuse-UDA outperforms leading UDA and semi-supervised strategies, achieving performance close to or even surpassing the theoretical upper bound of models trained directly on target domain data. Diffuse-UDA offers a pathway to advance the development and deployment of AI systems in medical imaging, addressing disparities between healthcare environments. This approach enables the exploration of innovative AI-driven diagnostic tools, improves outcomes, saves time, and reduces human error.
Intensity Confusion Matters: An Intensity-Distance Guided Loss for Bronchus Segmentation
Jun 23, 2024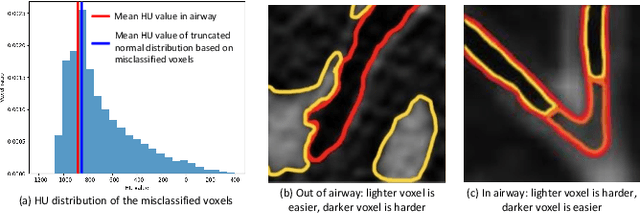
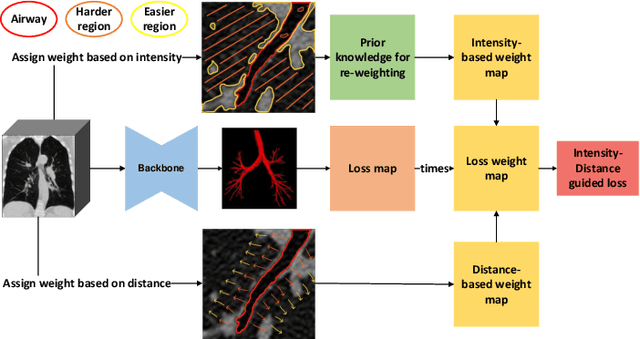
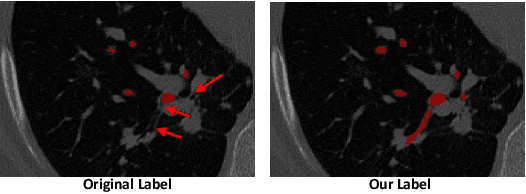
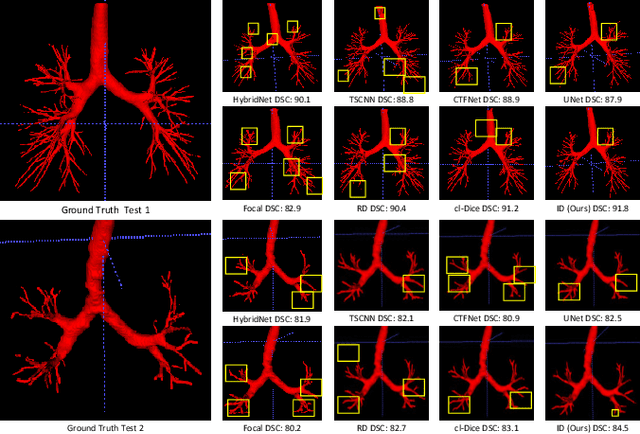
Abstract:Automatic segmentation of the bronchial tree from CT imaging is important, as it provides structural information for disease diagnosis. Despite the merits of previous automatic bronchus segmentation methods, they have paied less attention to the issue we term as \textit{Intensity Confusion}, wherein the intensity values of certain background voxels approach those of the foreground voxels within bronchi. Conversely, the intensity values of some foreground voxels are nearly identical to those of background voxels. This proximity in intensity values introduces significant challenges to neural network methodologies. To address the issue, we introduce a novel Intensity-Distance Guided loss function, which assigns adaptive weights to different image voxels for mining hard samples that cause the intensity confusion. The proposed loss estimates the voxel-level hardness of samples, on the basis of the following intensity and distance priors. We regard a voxel as a hard sample if it is in: (1) the background and has an intensity value close to the bronchus region; (2) the bronchus region and is of higher intensity than most voxels inside the bronchus; (3) the background region and at a short distance from the bronchus. Extensive experiments not only show the superiority of our method compared with the state-of-the-art methods, but also verify that tackling the intensity confusion issue helps to significantly improve bronchus segmentation. Project page: https://github.com/lhaof/ICM.
Self-Supervised Alignment Learning for Medical Image Segmentation
Jun 22, 2024Abstract:Recently, self-supervised learning (SSL) methods have been used in pre-training the segmentation models for 2D and 3D medical images. Most of these methods are based on reconstruction, contrastive learning and consistency regularization. However, the spatial correspondence of 2D slices from a 3D medical image has not been fully exploited. In this paper, we propose a novel self-supervised alignment learning framework to pre-train the neural network for medical image segmentation. The proposed framework consists of a new local alignment loss and a global positional loss. We observe that in the same 3D scan, two close 2D slices usually contain similar anatomic structures. Thus, the local alignment loss is proposed to make the pixel-level features of matched structures close to each other. Experimental results show that the proposed alignment learning is competitive with existing self-supervised pre-training approaches on CT and MRI datasets, under the setting of limited annotations.
Cell Graph Transformer for Nuclei Classification
Feb 20, 2024Abstract:Nuclei classification is a critical step in computer-aided diagnosis with histopathology images. In the past, various methods have employed graph neural networks (GNN) to analyze cell graphs that model inter-cell relationships by considering nuclei as vertices. However, they are limited by the GNN mechanism that only passes messages among local nodes via fixed edges. To address the issue, we develop a cell graph transformer (CGT) that treats nodes and edges as input tokens to enable learnable adjacency and information exchange among all nodes. Nevertheless, training the transformer with a cell graph presents another challenge. Poorly initialized features can lead to noisy self-attention scores and inferior convergence, particularly when processing the cell graphs with numerous connections. Thus, we further propose a novel topology-aware pretraining method that leverages a graph convolutional network (GCN) to learn a feature extractor. The pre-trained features may suppress unreasonable correlations and hence ease the finetuning of CGT. Experimental results suggest that the proposed cell graph transformer with topology-aware pretraining significantly improves the nuclei classification results, and achieves the state-of-the-art performance. Code and models are available at https://github.com/lhaof/CGT
UniCell: Universal Cell Nucleus Classification via Prompt Learning
Feb 20, 2024



Abstract:The recognition of multi-class cell nuclei can significantly facilitate the process of histopathological diagnosis. Numerous pathological datasets are currently available, but their annotations are inconsistent. Most existing methods require individual training on each dataset to deduce the relevant labels and lack the use of common knowledge across datasets, consequently restricting the quality of recognition. In this paper, we propose a universal cell nucleus classification framework (UniCell), which employs a novel prompt learning mechanism to uniformly predict the corresponding categories of pathological images from different dataset domains. In particular, our framework adopts an end-to-end architecture for nuclei detection and classification, and utilizes flexible prediction heads for adapting various datasets. Moreover, we develop a Dynamic Prompt Module (DPM) that exploits the properties of multiple datasets to enhance features. The DPM first integrates the embeddings of datasets and semantic categories, and then employs the integrated prompts to refine image representations, efficiently harvesting the shared knowledge among the related cell types and data sources. Experimental results demonstrate that the proposed method effectively achieves the state-of-the-art results on four nucleus detection and classification benchmarks. Code and models are available at https://github.com/lhaof/UniCell
nnMamba: 3D Biomedical Image Segmentation, Classification and Landmark Detection with State Space Model
Feb 05, 2024Abstract:In the field of biomedical image analysis, the quest for architectures capable of effectively capturing long-range dependencies is paramount, especially when dealing with 3D image segmentation, classification, and landmark detection. Traditional Convolutional Neural Networks (CNNs) struggle with locality respective field, and Transformers have a heavy computational load when applied to high-dimensional medical images. In this paper, we introduce nnMamba, a novel architecture that integrates the strengths of CNNs and the advanced long-range modeling capabilities of State Space Sequence Models (SSMs). nnMamba adds the SSMs to the convolutional residual-block to extract local features and model complex dependencies. For diffirent tasks, we build different blocks to learn the features. Extensive experiments demonstrate nnMamba's superiority over state-of-the-art methods in a suite of challenging tasks, including 3D image segmentation, classification, and landmark detection. nnMamba emerges as a robust solution, offering both the local representation ability of CNNs and the efficient global context processing of SSMs, setting a new standard for long-range dependency modeling in medical image analysis. Code is available at https://github.com/lhaof/nnMamba
Diffusion-based Data Augmentation for Nuclei Image Segmentation
Oct 22, 2023Abstract:Nuclei segmentation is a fundamental but challenging task in the quantitative analysis of histopathology images. Although fully-supervised deep learning-based methods have made significant progress, a large number of labeled images are required to achieve great segmentation performance. Considering that manually labeling all nuclei instances for a dataset is inefficient, obtaining a large-scale human-annotated dataset is time-consuming and labor-intensive. Therefore, augmenting a dataset with only a few labeled images to improve the segmentation performance is of significant research and application value. In this paper, we introduce the first diffusion-based augmentation method for nuclei segmentation. The idea is to synthesize a large number of labeled images to facilitate training the segmentation model. To achieve this, we propose a two-step strategy. In the first step, we train an unconditional diffusion model to synthesize the Nuclei Structure that is defined as the representation of pixel-level semantic and distance transform. Each synthetic nuclei structure will serve as a constraint on histopathology image synthesis and is further post-processed to be an instance map. In the second step, we train a conditioned diffusion model to synthesize histopathology images based on nuclei structures. The synthetic histopathology images paired with synthetic instance maps will be added to the real dataset for training the segmentation model. The experimental results show that by augmenting 10% labeled real dataset with synthetic samples, one can achieve comparable segmentation results with the fully-supervised baseline.
Prompt-based Grouping Transformer for Nucleus Detection and Classification
Oct 22, 2023Abstract:Automatic nuclei detection and classification can produce effective information for disease diagnosis. Most existing methods classify nuclei independently or do not make full use of the semantic similarity between nuclei and their grouping features. In this paper, we propose a novel end-to-end nuclei detection and classification framework based on a grouping transformer-based classifier. The nuclei classifier learns and updates the representations of nuclei groups and categories via hierarchically grouping the nucleus embeddings. Then the cell types are predicted with the pairwise correlations between categorical embeddings and nucleus features. For the efficiency of the fully transformer-based framework, we take the nucleus group embeddings as the input prompts of backbone, which helps harvest grouping guided features by tuning only the prompts instead of the whole backbone. Experimental results show that the proposed method significantly outperforms the existing models on three datasets.
Affine-Consistent Transformer for Multi-Class Cell Nuclei Detection
Oct 22, 2023



Abstract:Multi-class cell nuclei detection is a fundamental prerequisite in the diagnosis of histopathology. It is critical to efficiently locate and identify cells with diverse morphology and distributions in digital pathological images. Most existing methods take complex intermediate representations as learning targets and rely on inflexible post-refinements while paying less attention to various cell density and fields of view. In this paper, we propose a novel Affine-Consistent Transformer (AC-Former), which directly yields a sequence of nucleus positions and is trained collaboratively through two sub-networks, a global and a local network. The local branch learns to infer distorted input images of smaller scales while the global network outputs the large-scale predictions as extra supervision signals. We further introduce an Adaptive Affine Transformer (AAT) module, which can automatically learn the key spatial transformations to warp original images for local network training. The AAT module works by learning to capture the transformed image regions that are more valuable for training the model. Experimental results demonstrate that the proposed method significantly outperforms existing state-of-the-art algorithms on various benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge