Haifan Gong
Costal Cartilage Segmentation with Topology Guided Deformable Mamba: Method and Benchmark
Aug 14, 2024



Abstract:Costal cartilage segmentation is crucial to various medical applications, necessitating precise and reliable techniques due to its complex anatomy and the importance of accurate diagnosis and surgical planning. We propose a novel deep learning-based approach called topology-guided deformable Mamba (TGDM) for costal cartilage segmentation. The TGDM is tailored to capture the intricate long-range costal cartilage relationships. Our method leverages a deformable model that integrates topological priors to enhance the adaptability and accuracy of the segmentation process. Furthermore, we developed a comprehensive benchmark that contains 165 cases for costal cartilage segmentation. This benchmark sets a new standard for evaluating costal cartilage segmentation techniques and provides a valuable resource for future research. Extensive experiments conducted on both in-domain benchmarks and out-of domain test sets demonstrate the superiority of our approach over existing methods, showing significant improvements in segmentation precision and robustness.
Diffuse-UDA: Addressing Unsupervised Domain Adaptation in Medical Image Segmentation with Appearance and Structure Aligned Diffusion Models
Aug 12, 2024Abstract:The scarcity and complexity of voxel-level annotations in 3D medical imaging present significant challenges, particularly due to the domain gap between labeled datasets from well-resourced centers and unlabeled datasets from less-resourced centers. This disparity affects the fairness of artificial intelligence algorithms in healthcare. We introduce Diffuse-UDA, a novel method leveraging diffusion models to tackle Unsupervised Domain Adaptation (UDA) in medical image segmentation. Diffuse-UDA generates high-quality image-mask pairs with target domain characteristics and various structures, thereby enhancing UDA tasks. Initially, pseudo labels for target domain samples are generated. Subsequently, a specially tailored diffusion model, incorporating deformable augmentations, is trained on image-label or image-pseudo-label pairs from both domains. Finally, source domain labels guide the diffusion model to generate image-label pairs for the target domain. Comprehensive evaluations on several benchmarks demonstrate that Diffuse-UDA outperforms leading UDA and semi-supervised strategies, achieving performance close to or even surpassing the theoretical upper bound of models trained directly on target domain data. Diffuse-UDA offers a pathway to advance the development and deployment of AI systems in medical imaging, addressing disparities between healthcare environments. This approach enables the exploration of innovative AI-driven diagnostic tools, improves outcomes, saves time, and reduces human error.
Intensity Confusion Matters: An Intensity-Distance Guided Loss for Bronchus Segmentation
Jun 23, 2024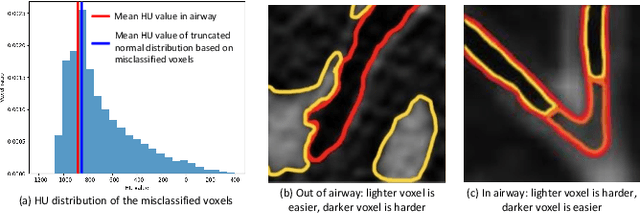
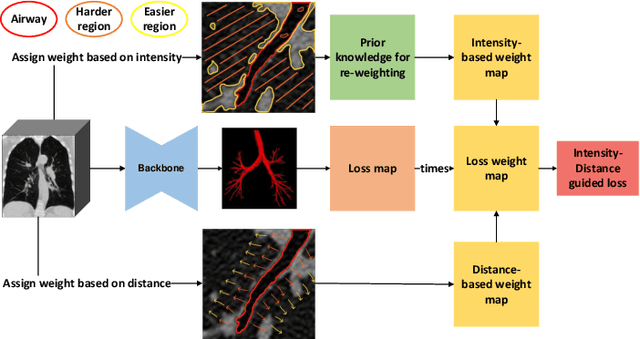
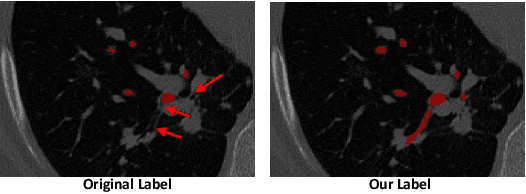
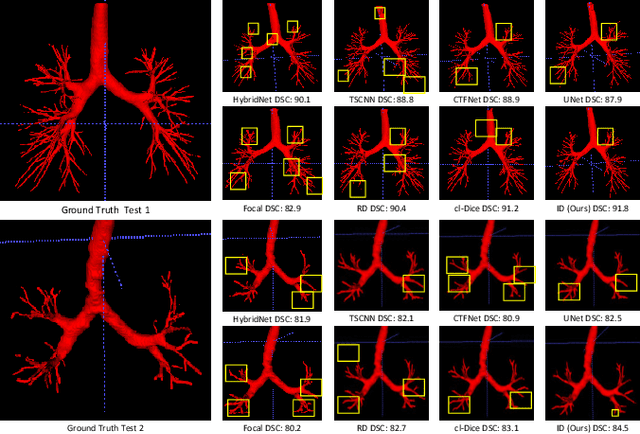
Abstract:Automatic segmentation of the bronchial tree from CT imaging is important, as it provides structural information for disease diagnosis. Despite the merits of previous automatic bronchus segmentation methods, they have paied less attention to the issue we term as \textit{Intensity Confusion}, wherein the intensity values of certain background voxels approach those of the foreground voxels within bronchi. Conversely, the intensity values of some foreground voxels are nearly identical to those of background voxels. This proximity in intensity values introduces significant challenges to neural network methodologies. To address the issue, we introduce a novel Intensity-Distance Guided loss function, which assigns adaptive weights to different image voxels for mining hard samples that cause the intensity confusion. The proposed loss estimates the voxel-level hardness of samples, on the basis of the following intensity and distance priors. We regard a voxel as a hard sample if it is in: (1) the background and has an intensity value close to the bronchus region; (2) the bronchus region and is of higher intensity than most voxels inside the bronchus; (3) the background region and at a short distance from the bronchus. Extensive experiments not only show the superiority of our method compared with the state-of-the-art methods, but also verify that tackling the intensity confusion issue helps to significantly improve bronchus segmentation. Project page: https://github.com/lhaof/ICM.
nnMamba: 3D Biomedical Image Segmentation, Classification and Landmark Detection with State Space Model
Feb 05, 2024Abstract:In the field of biomedical image analysis, the quest for architectures capable of effectively capturing long-range dependencies is paramount, especially when dealing with 3D image segmentation, classification, and landmark detection. Traditional Convolutional Neural Networks (CNNs) struggle with locality respective field, and Transformers have a heavy computational load when applied to high-dimensional medical images. In this paper, we introduce nnMamba, a novel architecture that integrates the strengths of CNNs and the advanced long-range modeling capabilities of State Space Sequence Models (SSMs). nnMamba adds the SSMs to the convolutional residual-block to extract local features and model complex dependencies. For diffirent tasks, we build different blocks to learn the features. Extensive experiments demonstrate nnMamba's superiority over state-of-the-art methods in a suite of challenging tasks, including 3D image segmentation, classification, and landmark detection. nnMamba emerges as a robust solution, offering both the local representation ability of CNNs and the efficient global context processing of SSMs, setting a new standard for long-range dependency modeling in medical image analysis. Code is available at https://github.com/lhaof/nnMamba
Visual-Attribute Prompt Learning for Progressive Mild Cognitive Impairment Prediction
Oct 22, 2023Abstract:Deep learning (DL) has been used in the automatic diagnosis of Mild Cognitive Impairment (MCI) and Alzheimer's Disease (AD) with brain imaging data. However, previous methods have not fully exploited the relation between brain image and clinical information that is widely adopted by experts in practice. To exploit the heterogeneous features from imaging and tabular data simultaneously, we propose the Visual-Attribute Prompt Learning-based Transformer (VAP-Former), a transformer-based network that efficiently extracts and fuses the multi-modal features with prompt fine-tuning. Furthermore, we propose a Prompt fine-Tuning (PT) scheme to transfer the knowledge from AD prediction task for progressive MCI (pMCI) diagnosis. In details, we first pre-train the VAP-Former without prompts on the AD diagnosis task and then fine-tune the model on the pMCI detection task with PT, which only needs to optimize a small amount of parameters while keeping the backbone frozen. Next, we propose a novel global prompt token for the visual prompts to provide global guidance to the multi-modal representations. Extensive experiments not only show the superiority of our method compared with the state-of-the-art methods in pMCI prediction but also demonstrate that the global prompt can make the prompt learning process more effective and stable. Interestingly, the proposed prompt learning model even outperforms the fully fine-tuning baseline on transferring the knowledge from AD to pMCI.
ASC: Appearance and Structure Consistency for Unsupervised Domain Adaptation in Fetal Brain MRI Segmentation
Oct 22, 2023Abstract:Automatic tissue segmentation of fetal brain images is essential for the quantitative analysis of prenatal neurodevelopment. However, producing voxel-level annotations of fetal brain imaging is time-consuming and expensive. To reduce labeling costs, we propose a practical unsupervised domain adaptation (UDA) setting that adapts the segmentation labels of high-quality fetal brain atlases to unlabeled fetal brain MRI data from another domain. To address the task, we propose a new UDA framework based on Appearance and Structure Consistency, named ASC. We adapt the segmentation model to the appearances of different domains by constraining the consistency before and after a frequency-based image transformation, which is to swap the appearance between brain MRI data and atlases. Consider that even in the same domain, the fetal brain images of different gestational ages could have significant variations in the anatomical structures. To make the model adapt to the structural variations in the target domain, we further encourage prediction consistency under different structural perturbations. Extensive experiments on FeTA 2021 benchmark demonstrate the effectiveness of our ASC in comparison to registration-based, semi-supervised learning-based, and existing UDA-based methods.
Less is More: Adaptive Curriculum Learning for Thyroid Nodule Diagnosis
Jul 02, 2022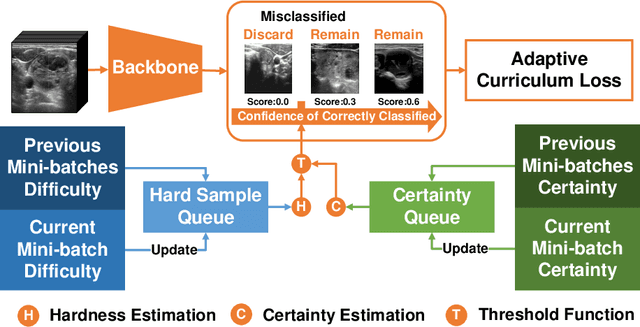
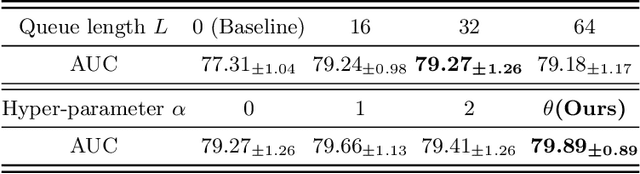
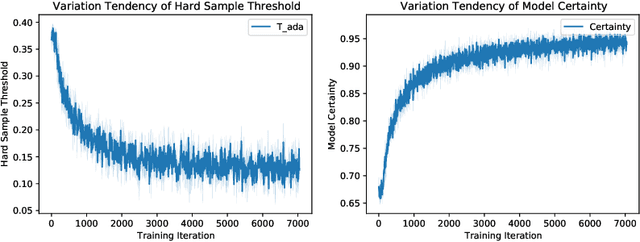
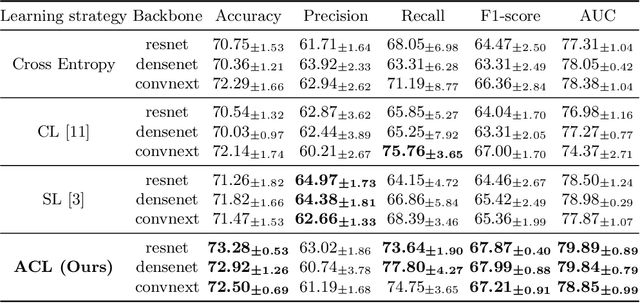
Abstract:Thyroid nodule classification aims at determining whether the nodule is benign or malignant based on a given ultrasound image. However, the label obtained by the cytological biopsy which is the golden standard in clinical medicine is not always consistent with the ultrasound imaging TI-RADS criteria. The information difference between the two causes the existing deep learning-based classification methods to be indecisive. To solve the Inconsistent Label problem, we propose an Adaptive Curriculum Learning (ACL) framework, which adaptively discovers and discards the samples with inconsistent labels. Specifically, ACL takes both hard sample and model certainty into account, and could accurately determine the threshold to distinguish the samples with Inconsistent Label. Moreover, we contribute TNCD: a Thyroid Nodule Classification Dataset to facilitate future related research on the thyroid nodules. Extensive experimental results on TNCD based on three different backbone networks not only demonstrate the superiority of our method but also prove that the less-is-more principle which strategically discards the samples with Inconsistent Label could yield performance gains. Source code and data are available at https://github.com/chenghui-666/ACL/.
BronchusNet: Region and Structure Prior Embedded Representation Learning for Bronchus Segmentation and Classification
May 24, 2022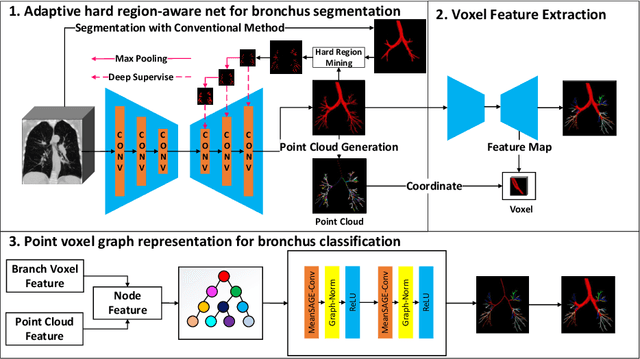



Abstract:CT-based bronchial tree analysis plays an important role in the computer-aided diagnosis for respiratory diseases, as it could provide structured information for clinicians. The basis of airway analysis is bronchial tree reconstruction, which consists of bronchus segmentation and classification. However, there remains a challenge for accurate bronchial analysis due to the individual variations and the severe class imbalance. In this paper, we propose a region and structure prior embedded framework named BronchusNet to achieve accurate segmentation and classification of bronchial regions in CT images. For bronchus segmentation, we propose an adaptive hard region-aware UNet that incorporates multi-level prior guidance of hard pixel-wise samples in the general Unet segmentation network to achieve better hierarchical feature learning. For the classification of bronchial branches, we propose a hybrid point-voxel graph learning module to fully exploit bronchial structure priors and to support simultaneous feature interactions across different branches. To facilitate the study of bronchial analysis, we contribute~\textbf{BRSC}: an open-access benchmark of \textbf{BR}onchus imaging analysis with high-quality pixel-wise \textbf{S}egmentation masks and the \textbf{C}lass of bronchial segments. Experimental results on BRSC show that our proposed method not only achieves the state-of-the-art performance for binary segmentation of bronchial region but also exceeds the best existing method on bronchial branches classification by 6.9\%.
A Survey of Natural Language Generation
Dec 22, 2021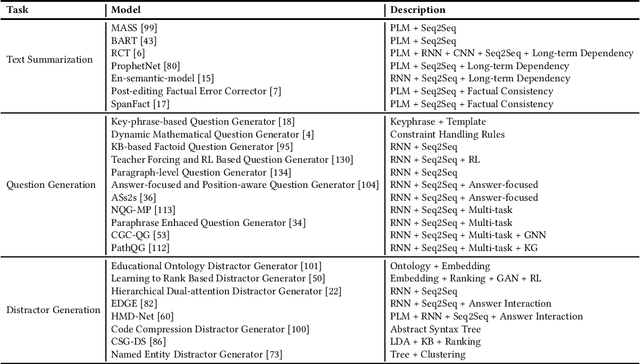
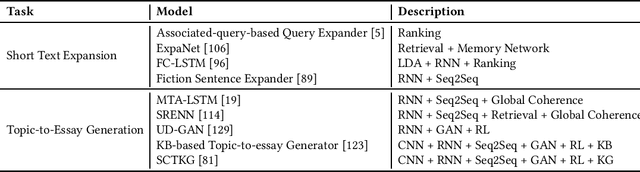
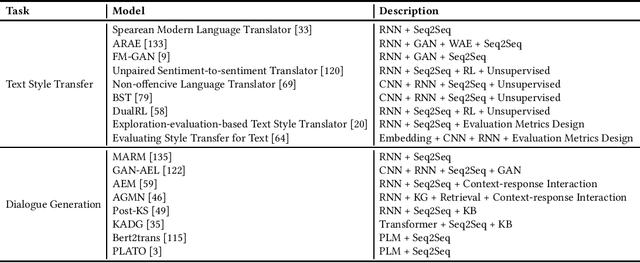
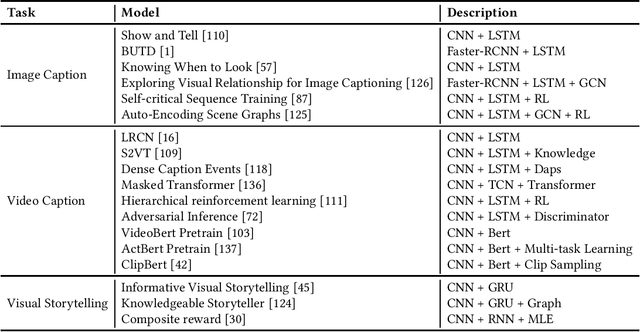
Abstract:This paper offers a comprehensive review of the research on Natural Language Generation (NLG) over the past two decades, especially in relation to data-to-text generation and text-to-text generation deep learning methods, as well as new applications of NLG technology. This survey aims to (a) give the latest synthesis of deep learning research on the NLG core tasks, as well as the architectures adopted in the field; (b) detail meticulously and comprehensively various NLG tasks and datasets, and draw attention to the challenges in NLG evaluation, focusing on different evaluation methods and their relationships; (c) highlight some future emphasis and relatively recent research issues that arise due to the increasing synergy between NLG and other artificial intelligence areas, such as computer vision, text and computational creativity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge