Xiaoying Lou
Neural Network-Based Histologic Remission Prediction In Ulcerative Colitis
Aug 28, 2023
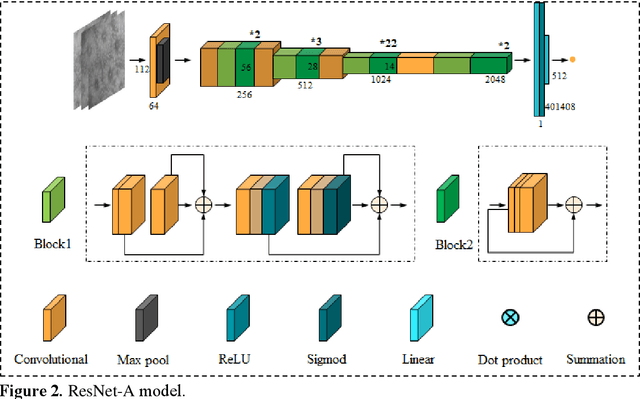
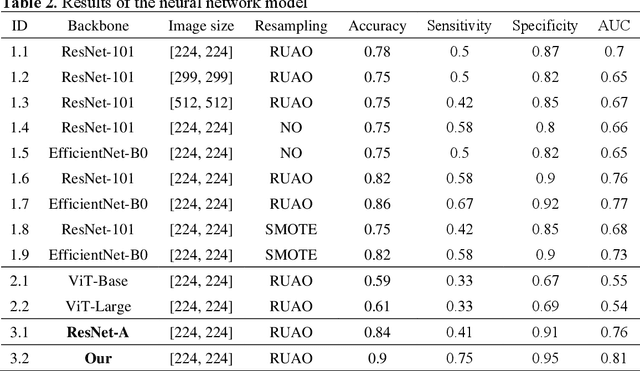
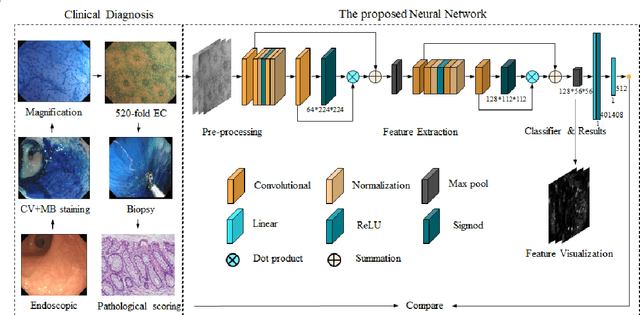
Abstract:BACKGROUND & AIMS: Histological remission (HR) is advocated and considered as a new therapeutic target in ulcerative colitis (UC). Diagnosis of histologic remission currently relies on biopsy; during this process, patients are at risk for bleeding, infection, and post-biopsy fibrosis. In addition, histologic response scoring is complex and time-consuming, and there is heterogeneity among pathologists. Endocytoscopy (EC) is a novel ultra-high magnification endoscopic technique that can provide excellent in vivo assessment of glands. Based on the EC technique, we propose a neural network model that can assess histological disease activity in UC using EC images to address the above issues. The experiment results demonstrate that the proposed method can assist patients in precise treatment and prognostic assessment. METHODS: We construct a neural network model for UC evaluation. A total of 5105 images of 154 intestinal segments from 87 patients undergoing EC treatment at a center in China between March 2022 and March 2023 are scored according to the Geboes score. Subsequently, 103 intestinal segments are used as the training set, 16 intestinal segments are used as the validation set for neural network training, and the remaining 35 intestinal segments are used as the test set to measure the model performance together with the validation set. RESULTS: By treating HR as a negative category and histologic activity as a positive category, the proposed neural network model can achieve an accuracy of 0.9, a specificity of 0.95, a sensitivity of 0.75, and an area under the curve (AUC) of 0.81. CONCLUSION: We develop a specific neural network model that can distinguish histologic remission/activity in EC images of UC, which helps to accelerate clinical histological diagnosis. keywords: ulcerative colitis; Endocytoscopy; Geboes score; neural network.
Structure Embedded Nucleus Classification for Histopathology Images
Feb 22, 2023Abstract:Nuclei classification provides valuable information for histopathology image analysis. However, the large variations in the appearance of different nuclei types cause difficulties in identifying nuclei. Most neural network based methods are affected by the local receptive field of convolutions, and pay less attention to the spatial distribution of nuclei or the irregular contour shape of a nucleus. In this paper, we first propose a novel polygon-structure feature learning mechanism that transforms a nucleus contour into a sequence of points sampled in order, and employ a recurrent neural network that aggregates the sequential change in distance between key points to obtain learnable shape features. Next, we convert a histopathology image into a graph structure with nuclei as nodes, and build a graph neural network to embed the spatial distribution of nuclei into their representations. To capture the correlations between the categories of nuclei and their surrounding tissue patterns, we further introduce edge features that are defined as the background textures between adjacent nuclei. Lastly, we integrate both polygon and graph structure learning mechanisms into a whole framework that can extract intra and inter-nucleus structural characteristics for nuclei classification. Experimental results show that the proposed framework achieves significant improvements compared to the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge