Tinglin Huang
Efficient Long-Horizon Vision-Language-Action Models via Static-Dynamic Disentanglement
Feb 03, 2026Abstract:Vision-Language-Action (VLA) models have recently emerged as a promising paradigm for generalist robotic control. Built upon vision-language model (VLM) architectures, VLAs predict actions conditioned on visual observations and language instructions, achieving strong performance and generalization across tasks. However, VLAs face two major challenges: limited long-horizon context and inefficient inference due to the quadratic attention complexity and large parameter counts. Our work is motivated by the observation that much of the visual information in a trajectory remains static across timesteps (e.g., the background). Leveraging this property, we propose SD-VLA, a framework that disentangles visual inputs into multi-level static and dynamic tokens, which enables (1) retaining a single copy of static tokens across frames to significantly reduce context length, and (2) reusing the key-value (KV) cache of static tokens through a lightweight recache gate that updates only when necessary. This design enables efficient multi-frame integration and efficient inference. In addition, we introduce a new benchmark that more effectively evaluates the long-horizon temporal dependency modeling ability of VLAs. Experimental results show that our approach outperforms baselines on this benchmark by 39.8% absolute improvement in success rate, and achieves a 3.9% gain on the SimplerEnv benchmark. Moreover, SD-VLA delivers a 2.26x inference speedup over the base VLA model on the same benchmark, enabling faster and more practical real-world deployment.
Patient-Similarity Cohort Reasoning in Clinical Text-to-SQL
Jan 14, 2026Abstract:Real-world clinical text-to-SQL requires reasoning over heterogeneous EHR tables, temporal windows, and patient-similarity cohorts to produce executable queries. We introduce CLINSQL, a benchmark of 633 expert-annotated tasks on MIMIC-IV v3.1 that demands multi-table joins, clinically meaningful filters, and executable SQL. Solving CLINSQL entails navigating schema metadata and clinical coding systems, handling long contexts, and composing multi-step queries beyond traditional text-to-SQL. We evaluate 22 proprietary and open-source models under Chain-of-Thought self-refinement and use rubric-based SQL analysis with execution checks that prioritize critical clinical requirements. Despite recent advances, performance remains far from clinical reliability: on the test set, GPT-5-mini attains 74.7% execution score, DeepSeek-R1 leads open-source at 69.2% and Gemini-2.5-Pro drops from 85.5% on Easy to 67.2% on Hard. Progress on CLINSQL marks tangible advances toward clinically reliable text-to-SQL for real-world EHR analytics.
HEIST: A Graph Foundation Model for Spatial Transcriptomics and Proteomics Data
Jun 11, 2025Abstract:Single-cell transcriptomics has become a great source for data-driven insights into biology, enabling the use of advanced deep learning methods to understand cellular heterogeneity and transcriptional regulation at the single-cell level. With the advent of spatial transcriptomics data we have the promise of learning about cells within a tissue context as it provides both spatial coordinates and transcriptomic readouts. However, existing models either ignore spatial resolution or the gene regulatory information. Gene regulation in cells can change depending on microenvironmental cues from neighboring cells, but existing models neglect gene regulatory patterns with hierarchical dependencies across levels of abstraction. In order to create contextualized representations of cells and genes from spatial transcriptomics data, we introduce HEIST, a hierarchical graph transformer-based foundation model for spatial transcriptomics and proteomics data. HEIST models tissue as spatial cellular neighborhood graphs, and each cell is, in turn, modeled as a gene regulatory network graph. The framework includes a hierarchical graph transformer that performs cross-level message passing and message passing within levels. HEIST is pre-trained on 22.3M cells from 124 tissues across 15 organs using spatially-aware contrastive learning and masked auto-encoding objectives. Unsupervised analysis of HEIST representations of cells, shows that it effectively encodes the microenvironmental influences in cell embeddings, enabling the discovery of spatially-informed subpopulations that prior models fail to differentiate. Further, HEIST achieves state-of-the-art results on four downstream task such as clinical outcome prediction, cell type annotation, gene imputation, and spatially-informed cell clustering across multiple technologies, highlighting the importance of hierarchical modeling and GRN-based representations.
Beyond Feature Importance: Feature Interactions in Predicting Post-Stroke Rigidity with Graph Explainable AI
Apr 10, 2025Abstract:This study addresses the challenge of predicting post-stroke rigidity by emphasizing feature interactions through graph-based explainable AI. Post-stroke rigidity, characterized by increased muscle tone and stiffness, significantly affects survivors' mobility and quality of life. Despite its prevalence, early prediction remains limited, delaying intervention. We analyze 519K stroke hospitalization records from the Healthcare Cost and Utilization Project dataset, where 43% of patients exhibited rigidity. We compare traditional approaches such as Logistic Regression, XGBoost, and Transformer with graph-based models like Graphormer and Graph Attention Network. These graph models inherently capture feature interactions and incorporate intrinsic or post-hoc explainability. Our results show that graph-based methods outperform others (AUROC 0.75), identifying key predictors such as NIH Stroke Scale and APR-DRG mortality risk scores. They also uncover interactions missed by conventional models. This research provides a novel application of graph-based XAI in stroke prognosis, with potential to guide early identification and personalized rehabilitation strategies.
SurfPro: Functional Protein Design Based on Continuous Surface
May 07, 2024Abstract:How can we design proteins with desired functions? We are motivated by a chemical intuition that both geometric structure and biochemical properties are critical to a protein's function. In this paper, we propose SurfPro, a new method to generate functional proteins given a desired surface and its associated biochemical properties. SurfPro comprises a hierarchical encoder that progressively models the geometric shape and biochemical features of a protein surface, and an autoregressive decoder to produce an amino acid sequence. We evaluate SurfPro on a standard inverse folding benchmark CATH 4.2 and two functional protein design tasks: protein binder design and enzyme design. Our SurfPro consistently surpasses previous state-of-the-art inverse folding methods, achieving a recovery rate of 57.78% on CATH 4.2 and higher success rates in terms of protein-protein binding and enzyme-substrate interaction scores.
Does Negative Sampling Matter? A Review with Insights into its Theory and Applications
Feb 27, 2024



Abstract:Negative sampling has swiftly risen to prominence as a focal point of research, with wide-ranging applications spanning machine learning, computer vision, natural language processing, data mining, and recommender systems. This growing interest raises several critical questions: Does negative sampling really matter? Is there a general framework that can incorporate all existing negative sampling methods? In what fields is it applied? Addressing these questions, we propose a general framework that leverages negative sampling. Delving into the history of negative sampling, we trace the development of negative sampling through five evolutionary paths. We dissect and categorize the strategies used to select negative sample candidates, detailing global, local, mini-batch, hop, and memory-based approaches. Our review categorizes current negative sampling methods into five types: static, hard, GAN-based, Auxiliary-based, and In-batch methods, providing a clear structure for understanding negative sampling. Beyond detailed categorization, we highlight the application of negative sampling in various areas, offering insights into its practical benefits. Finally, we briefly discuss open problems and future directions for negative sampling.
Learning to Group Auxiliary Datasets for Molecule
Jul 08, 2023Abstract:The limited availability of annotations in small molecule datasets presents a challenge to machine learning models. To address this, one common strategy is to collaborate with additional auxiliary datasets. However, having more data does not always guarantee improvements. Negative transfer can occur when the knowledge in the target dataset differs or contradicts that of the auxiliary molecule datasets. In light of this, identifying the auxiliary molecule datasets that can benefit the target dataset when jointly trained remains a critical and unresolved problem. Through an empirical analysis, we observe that combining graph structure similarity and task similarity can serve as a more reliable indicator for identifying high-affinity auxiliary datasets. Motivated by this insight, we propose MolGroup, which separates the dataset affinity into task and structure affinity to predict the potential benefits of each auxiliary molecule dataset. MolGroup achieves this by utilizing a routing mechanism optimized through a bi-level optimization framework. Empowered by the meta gradient, the routing mechanism is optimized toward maximizing the target dataset's performance and quantifies the affinity as the gating score. As a result, MolGroup is capable of predicting the optimal combination of auxiliary datasets for each target dataset. Our extensive experiments demonstrate the efficiency and effectiveness of MolGroup, showing an average improvement of 4.41%/3.47% for GIN/Graphormer trained with the group of molecule datasets selected by MolGroup on 11 target molecule datasets.
BatchSampler: Sampling Mini-Batches for Contrastive Learning in Vision, Language, and Graphs
Jun 06, 2023Abstract:In-Batch contrastive learning is a state-of-the-art self-supervised method that brings semantically-similar instances close while pushing dissimilar instances apart within a mini-batch. Its key to success is the negative sharing strategy, in which every instance serves as a negative for the others within the mini-batch. Recent studies aim to improve performance by sampling hard negatives \textit{within the current mini-batch}, whose quality is bounded by the mini-batch itself. In this work, we propose to improve contrastive learning by sampling mini-batches from the input data. We present BatchSampler\footnote{The code is available at \url{https://github.com/THUDM/BatchSampler}} to sample mini-batches of hard-to-distinguish (i.e., hard and true negatives to each other) instances. To make each mini-batch have fewer false negatives, we design the proximity graph of randomly-selected instances. To form the mini-batch, we leverage random walk with restart on the proximity graph to help sample hard-to-distinguish instances. BatchSampler is a simple and general technique that can be directly plugged into existing contrastive learning models in vision, language, and graphs. Extensive experiments on datasets of three modalities show that BatchSampler can consistently improve the performance of powerful contrastive models, as shown by significant improvements of SimCLR on ImageNet-100, SimCSE on STS (language), and GraphCL and MVGRL on graph datasets.
* 17 pages, 16 figures
GRAND+: Scalable Graph Random Neural Networks
Mar 12, 2022



Abstract:Graph neural networks (GNNs) have been widely adopted for semi-supervised learning on graphs. A recent study shows that the graph random neural network (GRAND) model can generate state-of-the-art performance for this problem. However, it is difficult for GRAND to handle large-scale graphs since its effectiveness relies on computationally expensive data augmentation procedures. In this work, we present a scalable and high-performance GNN framework GRAND+ for semi-supervised graph learning. To address the above issue, we develop a generalized forward push (GFPush) algorithm in GRAND+ to pre-compute a general propagation matrix and employ it to perform graph data augmentation in a mini-batch manner. We show that both the low time and space complexities of GFPush enable GRAND+ to efficiently scale to large graphs. Furthermore, we introduce a confidence-aware consistency loss into the model optimization of GRAND+, facilitating GRAND+'s generalization superiority. We conduct extensive experiments on seven public datasets of different sizes. The results demonstrate that GRAND+ 1) is able to scale to large graphs and costs less running time than existing scalable GNNs, and 2) can offer consistent accuracy improvements over both full-batch and scalable GNNs across all datasets.
Learning Intents behind Interactions with Knowledge Graph for Recommendation
Feb 14, 2021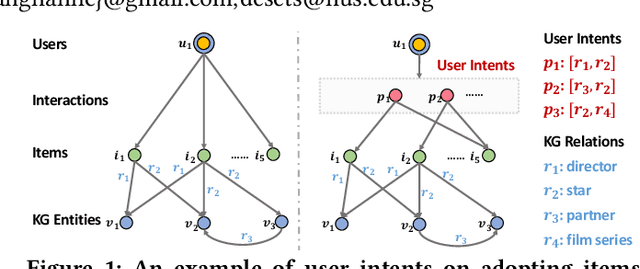

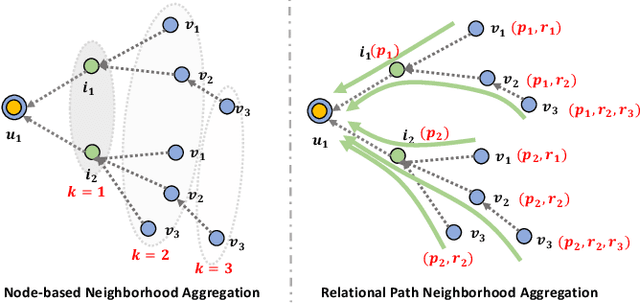

Abstract:Knowledge graph (KG) plays an increasingly important role in recommender systems. A recent technical trend is to develop end-to-end models founded on graph neural networks (GNNs). However, existing GNN-based models are coarse-grained in relational modeling, failing to (1) identify user-item relation at a fine-grained level of intents, and (2) exploit relation dependencies to preserve the semantics of long-range connectivity. In this study, we explore intents behind a user-item interaction by using auxiliary item knowledge, and propose a new model, Knowledge Graph-based Intent Network (KGIN). Technically, we model each intent as an attentive combination of KG relations, encouraging the independence of different intents for better model capability and interpretability. Furthermore, we devise a new information aggregation scheme for GNN, which recursively integrates the relation sequences of long-range connectivity (i.e., relational paths). This scheme allows us to distill useful information about user intents and encode them into the representations of users and items. Experimental results on three benchmark datasets show that, KGIN achieves significant improvements over the state-of-the-art methods like KGAT, KGNN-LS, and CKAN. Further analyses show that KGIN offers interpretable explanations for predictions by identifying influential intents and relational paths. The implementations are available at https://github.com/huangtinglin/Knowledge_Graph_based_Intent_Network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge