Stefan Hegselmann
Machine Learning for Health symposium 2024 -- Findings track
Mar 02, 2025Abstract:A collection of the accepted Findings papers that were presented at the 4th Machine Learning for Health symposium (ML4H 2024), which was held on December 15-16, 2024, in Vancouver, BC, Canada. ML4H 2024 invited high-quality submissions describing innovative research in a variety of health-related disciplines including healthcare, biomedicine, and public health. Works could be submitted to either the archival Proceedings track, or the non-archival Findings track. The Proceedings track targeted mature, cohesive works with technical sophistication and high-impact relevance to health. The Findings track promoted works that would spark new insights, collaborations, and discussions at ML4H. Both tracks were given the opportunity to share their work through the in-person poster session. All the manuscripts submitted to ML4H Symposium underwent a double-blind peer-review process.
Large Language Models are Powerful EHR Encoders
Feb 24, 2025Abstract:Electronic Health Records (EHRs) offer rich potential for clinical prediction, yet their inherent complexity and heterogeneity pose significant challenges for traditional machine learning approaches. Domain-specific EHR foundation models trained on large collections of unlabeled EHR data have demonstrated promising improvements in predictive accuracy and generalization; however, their training is constrained by limited access to diverse, high-quality datasets and inconsistencies in coding standards and healthcare practices. In this study, we explore the possibility of using general-purpose Large Language Models (LLMs) based embedding methods as EHR encoders. By serializing patient records into structured Markdown text, transforming codes into human-readable descriptors, we leverage the extensive generalization capabilities of LLMs pretrained on vast public corpora, thereby bypassing the need for proprietary medical datasets. We systematically evaluate two state-of-the-art LLM-embedding models, GTE-Qwen2-7B-Instruct and LLM2Vec-Llama3.1-8B-Instruct, across 15 diverse clinical prediction tasks from the EHRSHOT benchmark, comparing their performance to an EHRspecific foundation model, CLIMBR-T-Base, and traditional machine learning baselines. Our results demonstrate that LLM-based embeddings frequently match or exceed the performance of specialized models, even in few-shot settings, and that their effectiveness scales with the size of the underlying LLM and the available context window. Overall, our findings demonstrate that repurposing LLMs for EHR encoding offers a scalable and effective approach for clinical prediction, capable of overcoming the limitations of traditional EHR modeling and facilitating more interoperable and generalizable healthcare applications.
A Data-Centric Approach To Generate Faithful and High Quality Patient Summaries with Large Language Models
Feb 23, 2024



Abstract:Patients often face difficulties in understanding their hospitalizations, while healthcare workers have limited resources to provide explanations. In this work, we investigate the potential of large language models to generate patient summaries based on doctors' notes and study the effect of training data on the faithfulness and quality of the generated summaries. To this end, we develop a rigorous labeling protocol for hallucinations, and have two medical experts annotate 100 real-world summaries and 100 generated summaries. We show that fine-tuning on hallucination-free data effectively reduces hallucinations from 2.60 to 1.55 per summary for Llama 2, while preserving relevant information. Although the effect is still present, it is much smaller for GPT-4 when prompted with five examples (0.70 to 0.40). We also conduct a qualitative evaluation using hallucination-free and improved training data. GPT-4 shows very good results even in the zero-shot setting. We find that common quantitative metrics do not correlate well with faithfulness and quality. Finally, we test GPT-4 for automatic hallucination detection, which yields promising results.
Machine Learning for Health symposium 2023 -- Findings track
Dec 01, 2023Abstract:A collection of the accepted Findings papers that were presented at the 3rd Machine Learning for Health symposium (ML4H 2023), which was held on December 10, 2023, in New Orleans, Louisiana, USA. ML4H 2023 invited high-quality submissions on relevant problems in a variety of health-related disciplines including healthcare, biomedicine, and public health. Two submission tracks were offered: the archival Proceedings track, and the non-archival Findings track. Proceedings were targeted at mature work with strong technical sophistication and a high impact to health. The Findings track looked for new ideas that could spark insightful discussion, serve as valuable resources for the community, or could enable new collaborations. Submissions to the Proceedings track, if not accepted, were automatically considered for the Findings track. All the manuscripts submitted to ML4H Symposium underwent a double-blind peer-review process.
TabLLM: Few-shot Classification of Tabular Data with Large Language Models
Oct 19, 2022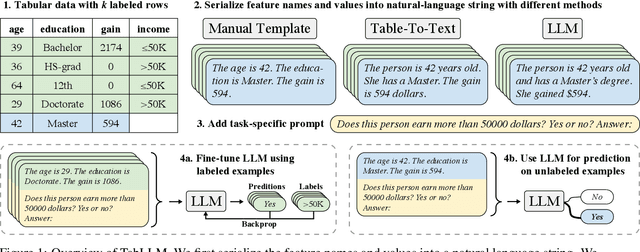
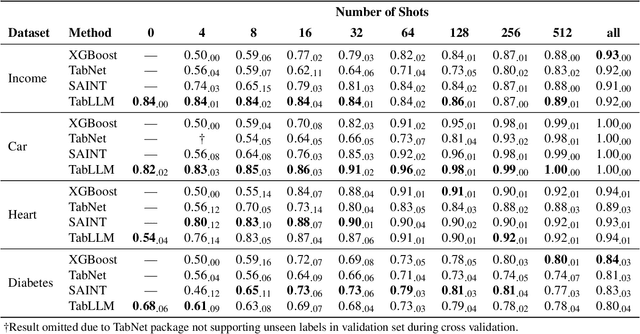
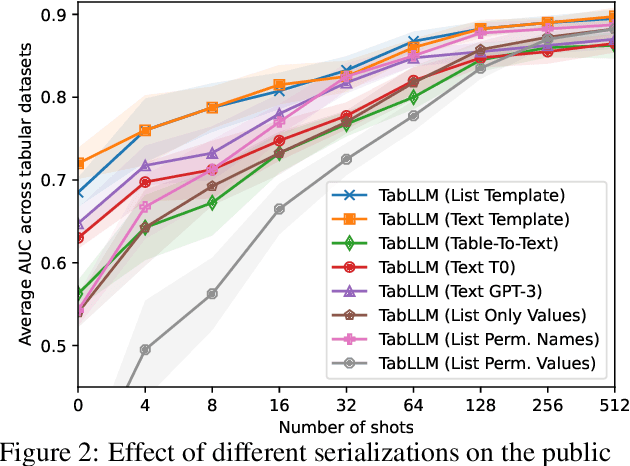
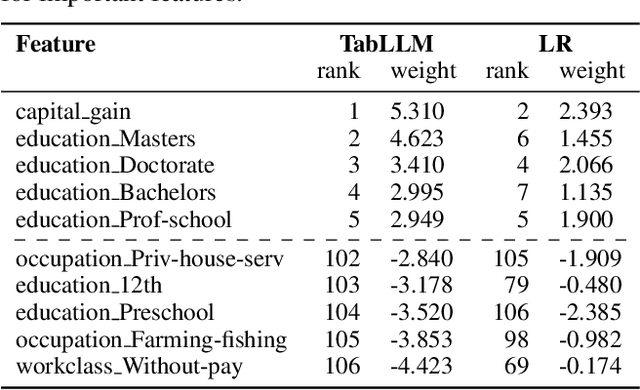
Abstract:We study the application of large language models to zero-shot and few-shot classification of tabular data. We prompt the large language model with a serialization of the tabular data to a natural-language string, together with a short description of the classification problem. In the few-shot setting, we fine-tune the large language model using some labeled examples. We evaluate several serialization methods including templates, table-to-text models, and large language models. Despite its simplicity, we find that this technique outperforms prior deep-learning-based tabular classification methods on several benchmark datasets. In most cases, even zero-shot classification obtains non-trivial performance, illustrating the method's ability to exploit prior knowledge encoded in large language models. Unlike many deep learning methods for tabular datasets, this approach is also competitive with strong traditional baselines like gradient-boosted trees, especially in the very-few-shot setting.
Large Language Models are Zero-Shot Clinical Information Extractors
May 25, 2022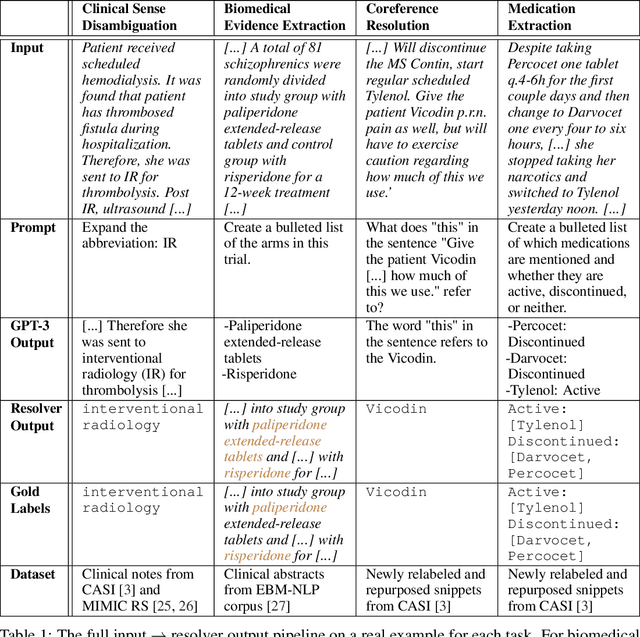
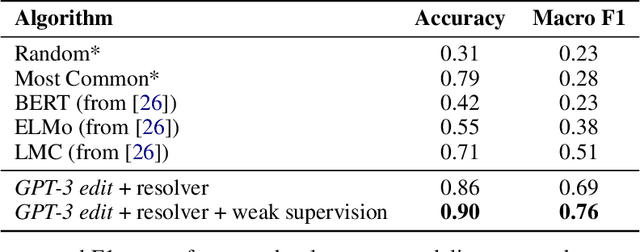

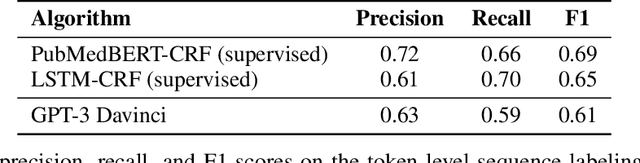
Abstract:We show that large language models, such as GPT-3, perform well at zero-shot information extraction from clinical text despite not being trained specifically for the clinical domain. We present several examples showing how to use these models as tools for the diverse tasks of (i) concept disambiguation, (ii) evidence extraction, (iii) coreference resolution, and (iv) concept extraction, all on clinical text. The key to good performance is the use of simple task-specific programs that map from the language model outputs to the label space of the task. We refer to these programs as resolvers, a generalization of the verbalizer, which defines a mapping between output tokens and a discrete label space. We show in our examples that good resolvers share common components (e.g., "safety checks" that ensure the language model outputs faithfully match the input data), and that the common patterns across tasks make resolvers lightweight and easy to create. To better evaluate these systems, we also introduce two new datasets for benchmarking zero-shot clinical information extraction based on manual relabeling of the CASI dataset (Moon et al., 2014) with labels for new tasks. On the clinical extraction tasks we studied, the GPT-3 + resolver systems significantly outperform existing zero- and few-shot baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge