Large Language Models are Zero-Shot Clinical Information Extractors
Paper and Code
May 25, 2022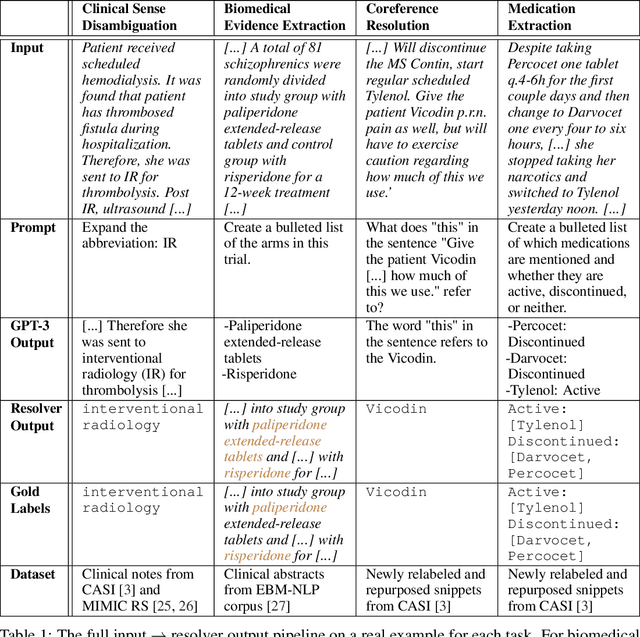
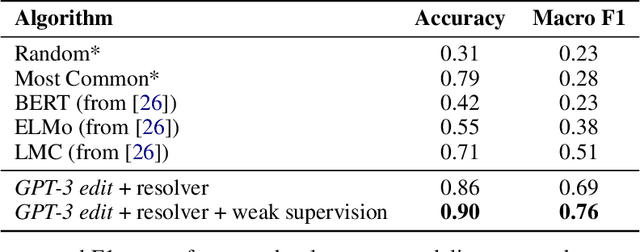

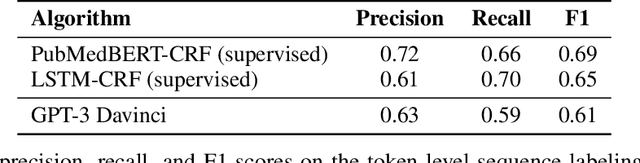
We show that large language models, such as GPT-3, perform well at zero-shot information extraction from clinical text despite not being trained specifically for the clinical domain. We present several examples showing how to use these models as tools for the diverse tasks of (i) concept disambiguation, (ii) evidence extraction, (iii) coreference resolution, and (iv) concept extraction, all on clinical text. The key to good performance is the use of simple task-specific programs that map from the language model outputs to the label space of the task. We refer to these programs as resolvers, a generalization of the verbalizer, which defines a mapping between output tokens and a discrete label space. We show in our examples that good resolvers share common components (e.g., "safety checks" that ensure the language model outputs faithfully match the input data), and that the common patterns across tasks make resolvers lightweight and easy to create. To better evaluate these systems, we also introduce two new datasets for benchmarking zero-shot clinical information extraction based on manual relabeling of the CASI dataset (Moon et al., 2014) with labels for new tasks. On the clinical extraction tasks we studied, the GPT-3 + resolver systems significantly outperform existing zero- and few-shot baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge