Tillmann Rheude
Fusion or Confusion? Multimodal Complexity Is Not All You Need
Dec 28, 2025Abstract:Deep learning architectures for multimodal learning have increased in complexity, driven by the assumption that multimodal-specific methods improve performance. We challenge this assumption through a large-scale empirical study reimplementing 19 high-impact methods under standardized conditions, evaluating them across nine diverse datasets with up to 23 modalities, and testing their generalizability to new tasks beyond their original scope, including settings with missing modalities. We propose a Simple Baseline for Multimodal Learning (SimBaMM), a straightforward late-fusion Transformer architecture, and demonstrate that under standardized experimental conditions with rigorous hyperparameter tuning of all methods, more complex architectures do not reliably outperform SimBaMM. Statistical analysis indicates that more complex methods perform comparably to SimBaMM and frequently do not reliably outperform well-tuned unimodal baselines, especially in the small-data regime considered in many original studies. To support our findings, we include a case study of a recent multimodal learning method highlighting the methodological shortcomings in the literature. In addition, we provide a pragmatic reliability checklist to promote comparable, robust, and trustworthy future evaluations. In summary, we argue for a shift in focus: away from the pursuit of architectural novelty and toward methodological rigor.
Cohort-Based Active Modality Acquisition
May 22, 2025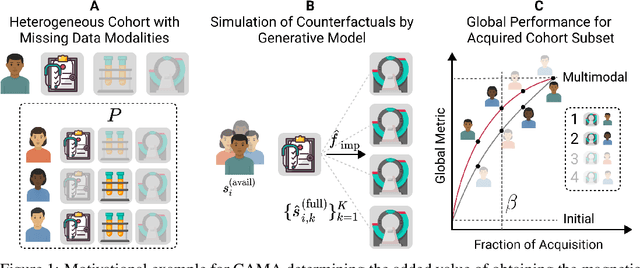
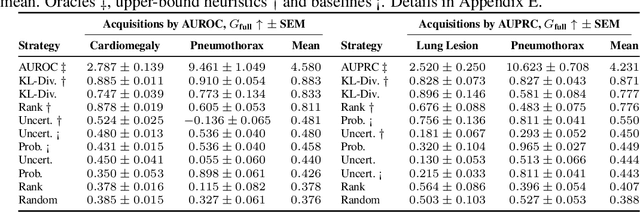
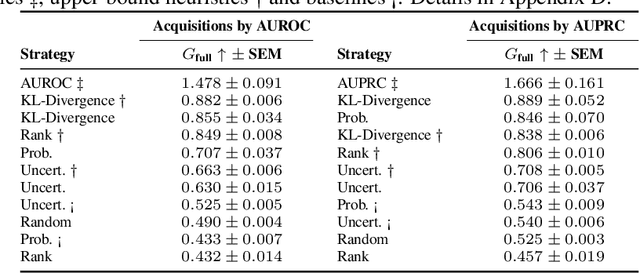
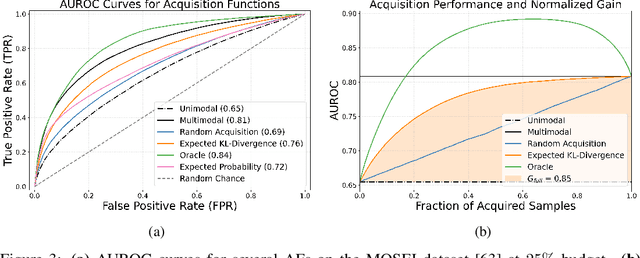
Abstract:Real-world machine learning applications often involve data from multiple modalities that must be integrated effectively to make robust predictions. However, in many practical settings, not all modalities are available for every sample, and acquiring additional modalities can be costly. This raises the question: which samples should be prioritized for additional modality acquisition when resources are limited? While prior work has explored individual-level acquisition strategies and training-time active learning paradigms, test-time and cohort-based acquisition remain underexplored despite their importance in many real-world settings. We introduce Cohort-based Active Modality Acquisition (CAMA), a novel test-time setting to formalize the challenge of selecting which samples should receive additional modalities. We derive acquisition strategies that leverage a combination of generative imputation and discriminative modeling to estimate the expected benefit of acquiring missing modalities based on common evaluation metrics. We also introduce upper-bound heuristics that provide performance ceilings to benchmark acquisition strategies. Experiments on common multimodal datasets demonstrate that our proposed imputation-based strategies can more effectively guide the acquisition of new samples in comparison to those relying solely on unimodal information, entropy guidance, and random selections. Our work provides an effective solution for optimizing modality acquisition at the cohort level, enabling better utilization of resources in constrained settings.
Large Language Models are Powerful EHR Encoders
Feb 24, 2025Abstract:Electronic Health Records (EHRs) offer rich potential for clinical prediction, yet their inherent complexity and heterogeneity pose significant challenges for traditional machine learning approaches. Domain-specific EHR foundation models trained on large collections of unlabeled EHR data have demonstrated promising improvements in predictive accuracy and generalization; however, their training is constrained by limited access to diverse, high-quality datasets and inconsistencies in coding standards and healthcare practices. In this study, we explore the possibility of using general-purpose Large Language Models (LLMs) based embedding methods as EHR encoders. By serializing patient records into structured Markdown text, transforming codes into human-readable descriptors, we leverage the extensive generalization capabilities of LLMs pretrained on vast public corpora, thereby bypassing the need for proprietary medical datasets. We systematically evaluate two state-of-the-art LLM-embedding models, GTE-Qwen2-7B-Instruct and LLM2Vec-Llama3.1-8B-Instruct, across 15 diverse clinical prediction tasks from the EHRSHOT benchmark, comparing their performance to an EHRspecific foundation model, CLIMBR-T-Base, and traditional machine learning baselines. Our results demonstrate that LLM-based embeddings frequently match or exceed the performance of specialized models, even in few-shot settings, and that their effectiveness scales with the size of the underlying LLM and the available context window. Overall, our findings demonstrate that repurposing LLMs for EHR encoding offers a scalable and effective approach for clinical prediction, capable of overcoming the limitations of traditional EHR modeling and facilitating more interoperable and generalizable healthcare applications.
Leveraging CAM Algorithms for Explaining Medical Semantic Segmentation
Sep 30, 2024Abstract:Convolutional neural networks (CNNs) achieve prevailing results in segmentation tasks nowadays and represent the state-of-the-art for image-based analysis. However, the understanding of the accurate decision-making process of a CNN is rather unknown. The research area of explainable artificial intelligence (xAI) primarily revolves around understanding and interpreting this black-box behavior. One way of interpreting a CNN is the use of class activation maps (CAMs) that represent heatmaps to indicate the importance of image areas for the prediction of the CNN. For classification tasks, a variety of CAM algorithms exist. But for segmentation tasks, only one CAM algorithm for the interpretation of the output of a CNN exist. We propose a transfer between existing classification- and segmentation-based methods for more detailed, explainable, and consistent results which show salient pixels in semantic segmentation tasks. The resulting Seg-HiRes-Grad CAM is an extension of the segmentation-based Seg-Grad CAM with the transfer to the classification-based HiRes CAM. Our method improves the previously-mentioned existing segmentation-based method by adjusting it to recently published classification-based methods. Especially for medical image segmentation, this transfer solves existing explainability disadvantages.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:023
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge