Sofia Ira Ktena
Deep Bayesian Bandits: Exploring in Online Personalized Recommendations
Aug 03, 2020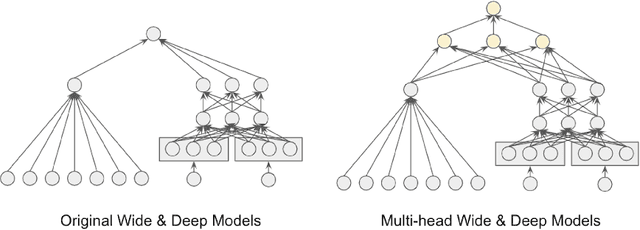
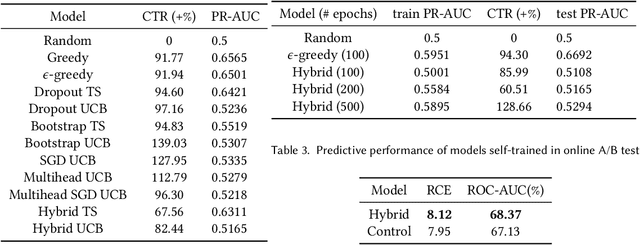
Abstract:Recommender systems trained in a continuous learning fashion are plagued by the feedback loop problem, also known as algorithmic bias. This causes a newly trained model to act greedily and favor items that have already been engaged by users. This behavior is particularly harmful in personalised ads recommendations, as it can also cause new campaigns to remain unexplored. Exploration aims to address this limitation by providing new information about the environment, which encompasses user preference, and can lead to higher long-term reward. In this work, we formulate a display advertising recommender as a contextual bandit and implement exploration techniques that require sampling from the posterior distribution of click-through-rates in a computationally tractable manner. Traditional large-scale deep learning models do not provide uncertainty estimates by default. We approximate these uncertainty measurements of the predictions by employing a bootstrapped model with multiple heads and dropout units. We benchmark a number of different models in an offline simulation environment using a publicly available dataset of user-ads engagements. We test our proposed deep Bayesian bandits algorithm in the offline simulation and online AB setting with large-scale production traffic, where we demonstrate a positive gain of our exploration model.
Model Size Reduction Using Frequency Based Double Hashing for Recommender Systems
Jul 28, 2020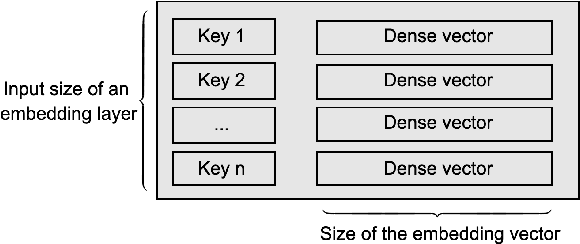
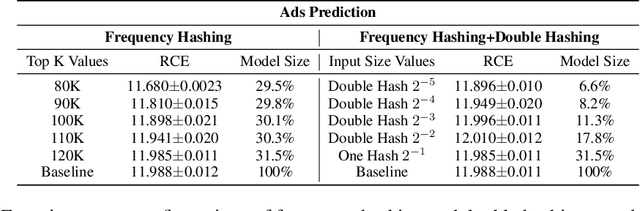
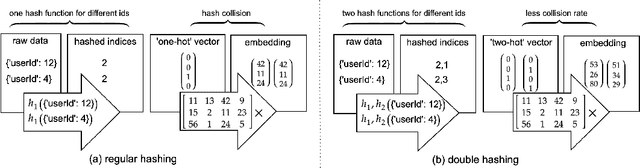
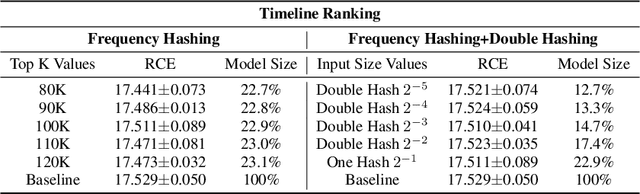
Abstract:Deep Neural Networks (DNNs) with sparse input features have been widely used in recommender systems in industry. These models have large memory requirements and need a huge amount of training data. The large model size usually entails a cost, in the range of millions of dollars, for storage and communication with the inference services. In this paper, we propose a hybrid hashing method to combine frequency hashing and double hashing techniques for model size reduction, without compromising performance. We evaluate the proposed models on two product surfaces. In both cases, experiment results demonstrated that we can reduce the model size by around 90 % while keeping the performance on par with the original baselines.
Privacy-Preserving Recommender Systems Challenge on Twitter's Home Timeline
Apr 28, 2020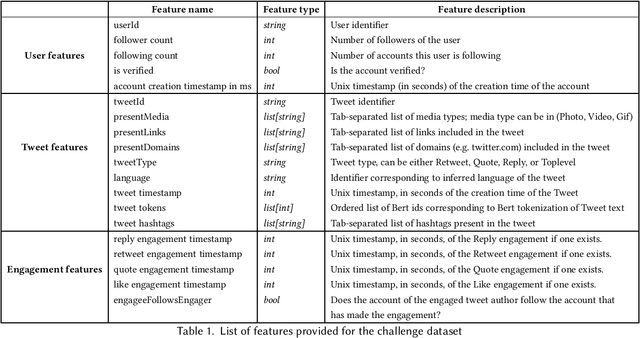
Abstract:Recommender systems constitute the core engine of most social network platforms nowadays, aiming to maximize user satisfaction along with other key business objectives. Twitter is no exception. Despite the fact that Twitter data has been extensively used to understand socioeconomic and political phenomena and user behaviour, the implicit feedback provided by users on Tweets through their engagements on the Home Timeline has only been explored to a limited extent. At the same time, there is a lack of large-scale public social network datasets that would enable the scientific community to both benchmark and build more powerful and comprehensive models that tailor content to user interests. By releasing an original dataset of 160 million Tweets along with engagement information, Twitter aims to address exactly that. During this release, special attention is drawn on maintaining compliance with existing privacy laws. Apart from user privacy, this paper touches on the key challenges faced by researchers and professionals striving to predict user engagements. It further describes the key aspects of the RecSys 2020 Challenge that was organized by ACM RecSys in partnership with Twitter using this dataset.
Addressing Delayed Feedback for Continuous Training with Neural Networks in CTR prediction
Jul 15, 2019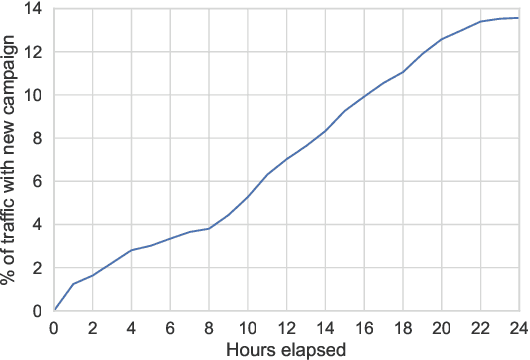
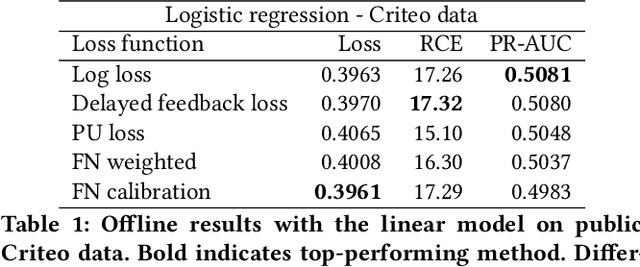
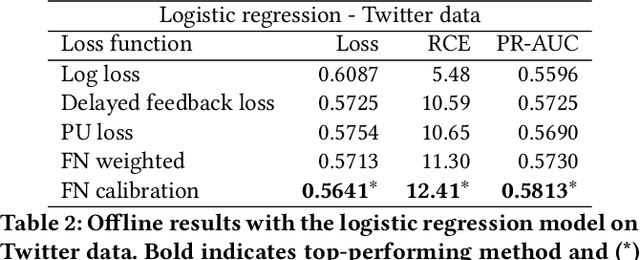
Abstract:One of the challenges in display advertising is that the distribution of features and click through rate (CTR) can exhibit large shifts over time due to seasonality, changes to ad campaigns and other factors. The predominant strategy to keep up with these shifts is to train predictive models continuously, on fresh data, in order to prevent them from becoming stale. However, in many ad systems positive labels are only observed after a possibly long and random delay. These delayed labels pose a challenge to data freshness in continuous training: fresh data may not have complete label information at the time they are ingested by the training algorithm. Naive strategies which consider any data point a negative example until a positive label becomes available tend to underestimate CTR, resulting in inferior user experience and suboptimal performance for advertisers. The focus of this paper is to identify the best combination of loss functions and models that enable large-scale learning from a continuous stream of data in the presence of delayed labels. In this work, we compare 5 different loss functions, 3 of them applied to this problem for the first time. We benchmark their performance in offline settings on both public and proprietary datasets in conjunction with shallow and deep model architectures. We also discuss the engineering cost associated with implementing each loss function in a production environment. Finally, we carried out online experiments with the top performing methods, in order to validate their performance in a continuous training scheme. While training on 668 million in-house data points offline, our proposed methods outperform previous state-of-the-art by 3% relative cross entropy (RCE). During online experiments, we observed 55% gain in revenue per thousand requests (RPMq) against naive log loss.
Graph Saliency Maps through Spectral Convolutional Networks: Application to Sex Classification with Brain Connectivity
Jun 05, 2018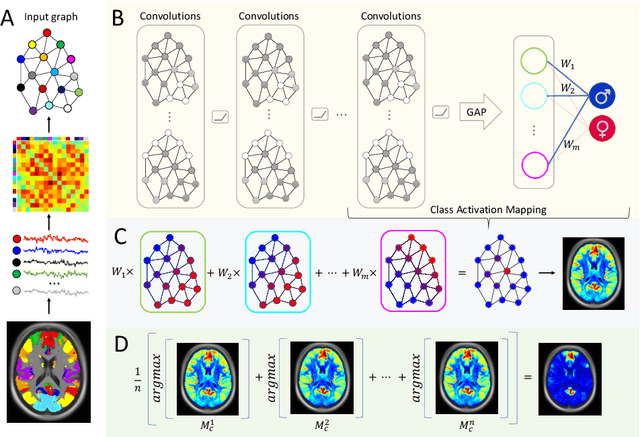
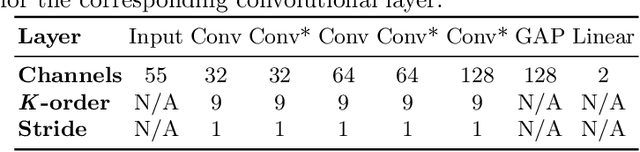
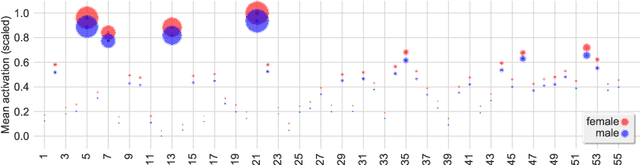
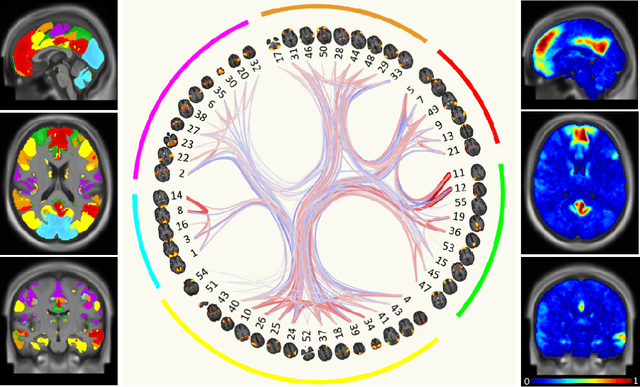
Abstract:Graph convolutional networks (GCNs) allow to apply traditional convolution operations in non-Euclidean domains, where data are commonly modelled as irregular graphs. Medical imaging and, in particular, neuroscience studies often rely on such graph representations, with brain connectivity networks being a characteristic example, while ultimately seeking the locus of phenotypic or disease-related differences in the brain. These regions of interest (ROIs) are, then, considered to be closely associated with function and/or behaviour. Driven by this, we explore GCNs for the task of ROI identification and propose a visual attribution method based on class activation mapping. By undertaking a sex classification task as proof of concept, we show that this method can be used to identify salient nodes (brain regions) without prior node labels. Based on experiments conducted on neuroimaging data of more than 5000 participants from UK Biobank, we demonstrate the robustness of the proposed method in highlighting reproducible regions across individuals. We further evaluate the neurobiological relevance of the identified regions based on evidence from large-scale UK Biobank studies.
Disease Prediction using Graph Convolutional Networks: Application to Autism Spectrum Disorder and Alzheimer's Disease
Jun 05, 2018



Abstract:Graphs are widely used as a natural framework that captures interactions between individual elements represented as nodes in a graph. In medical applications, specifically, nodes can represent individuals within a potentially large population (patients or healthy controls) accompanied by a set of features, while the graph edges incorporate associations between subjects in an intuitive manner. This representation allows to incorporate the wealth of imaging and non-imaging information as well as individual subject features simultaneously in disease classification tasks. Previous graph-based approaches for supervised or unsupervised learning in the context of disease prediction solely focus on pairwise similarities between subjects, disregarding individual characteristics and features, or rather rely on subject-specific imaging feature vectors and fail to model interactions between them. In this paper, we present a thorough evaluation of a generic framework that leverages both imaging and non-imaging information and can be used for brain analysis in large populations. This framework exploits Graph Convolutional Networks (GCNs) and involves representing populations as a sparse graph, where its nodes are associated with imaging-based feature vectors, while phenotypic information is integrated as edge weights. The extensive evaluation explores the effect of each individual component of this framework on disease prediction performance and further compares it to different baselines. The framework performance is tested on two large datasets with diverse underlying data, ABIDE and ADNI, for the prediction of Autism Spectrum Disorder and conversion to Alzheimer's disease, respectively. Our analysis shows that our novel framework can improve over state-of-the-art results on both databases, with 70.4% classification accuracy for ABIDE and 80.0% for ADNI.
DLTK: State of the Art Reference Implementations for Deep Learning on Medical Images
Nov 18, 2017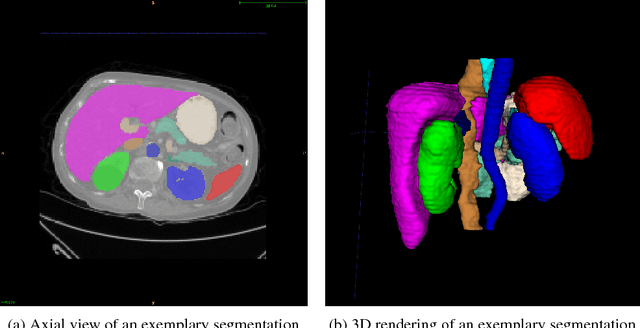
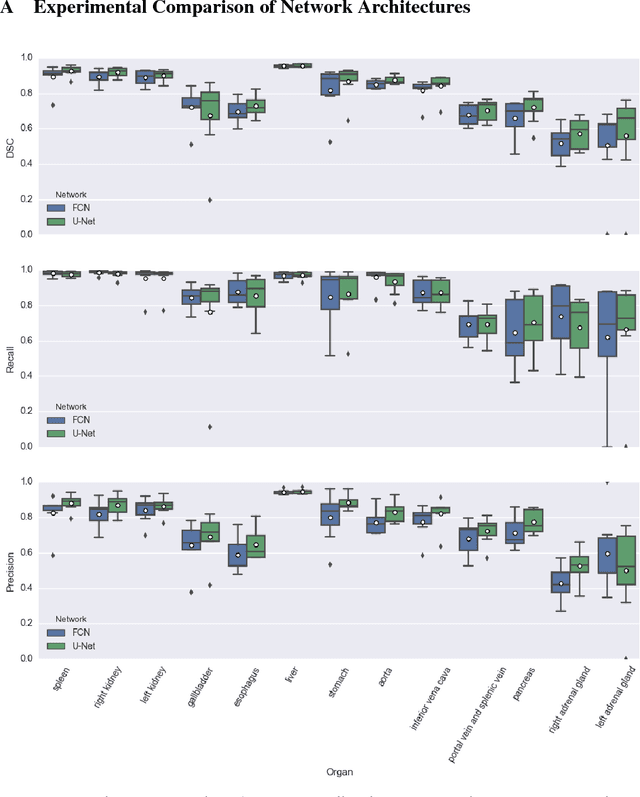
Abstract:We present DLTK, a toolkit providing baseline implementations for efficient experimentation with deep learning methods on biomedical images. It builds on top of TensorFlow and its high modularity and easy-to-use examples allow for a low-threshold access to state-of-the-art implementations for typical medical imaging problems. A comparison of DLTK's reference implementations of popular network architectures for image segmentation demonstrates new top performance on the publicly available challenge data "Multi-Atlas Labeling Beyond the Cranial Vault". The average test Dice similarity coefficient of $81.5$ exceeds the previously best performing CNN ($75.7$) and the accuracy of the challenge winning method ($79.0$).
Spectral Graph Convolutions for Population-based Disease Prediction
Jun 21, 2017

Abstract:Exploiting the wealth of imaging and non-imaging information for disease prediction tasks requires models capable of representing, at the same time, individual features as well as data associations between subjects from potentially large populations. Graphs provide a natural framework for such tasks, yet previous graph-based approaches focus on pairwise similarities without modelling the subjects' individual characteristics and features. On the other hand, relying solely on subject-specific imaging feature vectors fails to model the interaction and similarity between subjects, which can reduce performance. In this paper, we introduce the novel concept of Graph Convolutional Networks (GCN) for brain analysis in populations, combining imaging and non-imaging data. We represent populations as a sparse graph where its vertices are associated with image-based feature vectors and the edges encode phenotypic information. This structure was used to train a GCN model on partially labelled graphs, aiming to infer the classes of unlabelled nodes from the node features and pairwise associations between subjects. We demonstrate the potential of the method on the challenging ADNI and ABIDE databases, as a proof of concept of the benefit from integrating contextual information in classification tasks. This has a clear impact on the quality of the predictions, leading to 69.5% accuracy for ABIDE (outperforming the current state of the art of 66.8%) and 77% for ADNI for prediction of MCI conversion, significantly outperforming standard linear classifiers where only individual features are considered.
Distance Metric Learning using Graph Convolutional Networks: Application to Functional Brain Networks
Jun 14, 2017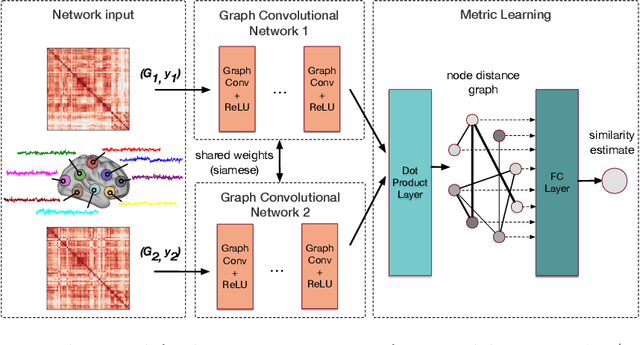

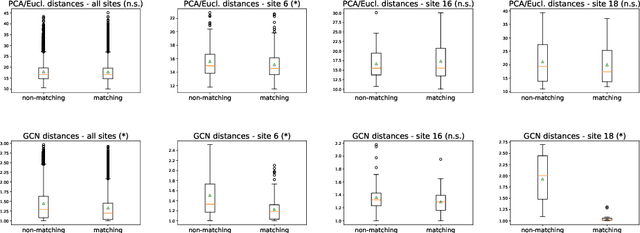
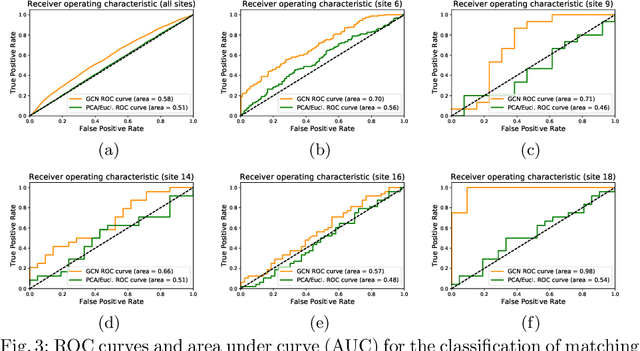
Abstract:Evaluating similarity between graphs is of major importance in several computer vision and pattern recognition problems, where graph representations are often used to model objects or interactions between elements. The choice of a distance or similarity metric is, however, not trivial and can be highly dependent on the application at hand. In this work, we propose a novel metric learning method to evaluate distance between graphs that leverages the power of convolutional neural networks, while exploiting concepts from spectral graph theory to allow these operations on irregular graphs. We demonstrate the potential of our method in the field of connectomics, where neuronal pathways or functional connections between brain regions are commonly modelled as graphs. In this problem, the definition of an appropriate graph similarity function is critical to unveil patterns of disruptions associated with certain brain disorders. Experimental results on the ABIDE dataset show that our method can learn a graph similarity metric tailored for a clinical application, improving the performance of a simple k-nn classifier by 11.9% compared to a traditional distance metric.
Exploring Heritability of Functional Brain Networks with Inexact Graph Matching
Mar 29, 2017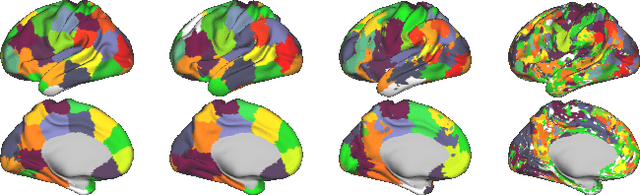
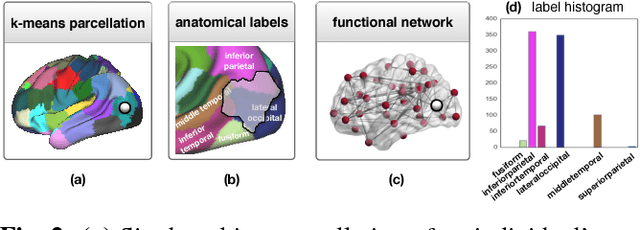
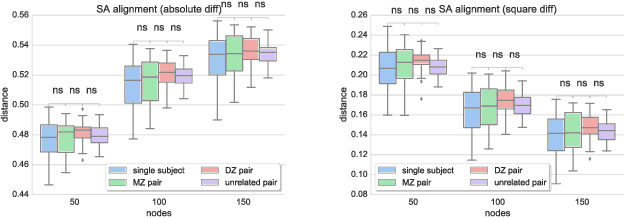
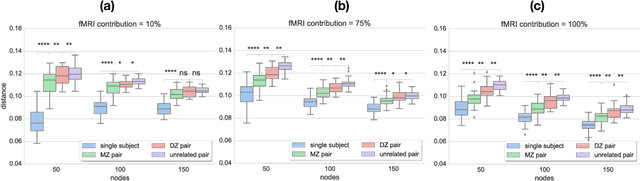
Abstract:Data-driven brain parcellations aim to provide a more accurate representation of an individual's functional connectivity, since they are able to capture individual variability that arises due to development or disease. This renders comparisons between the emerging brain connectivity networks more challenging, since correspondences between their elements are not preserved. Unveiling these correspondences is of major importance to keep track of local functional connectivity changes. We propose a novel method based on graph edit distance for the comparison of brain graphs directly in their domain, that can accurately reflect similarities between individual networks while providing the network element correspondences. This method is validated on a dataset of 116 twin subjects provided by the Human Connectome Project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge