Graph Saliency Maps through Spectral Convolutional Networks: Application to Sex Classification with Brain Connectivity
Paper and Code
Jun 05, 2018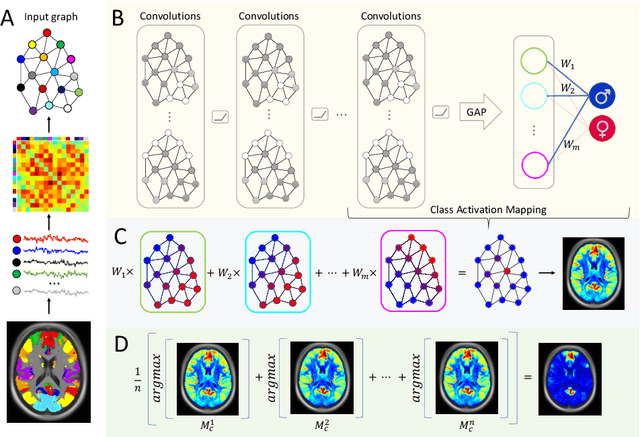
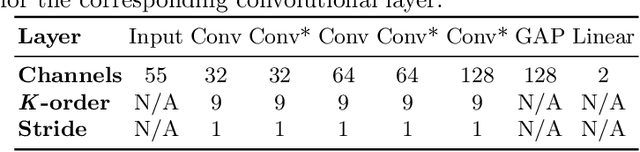
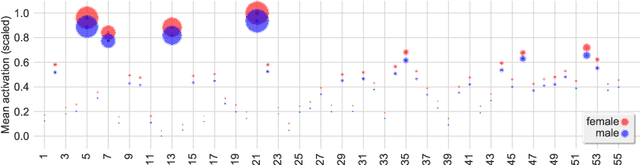
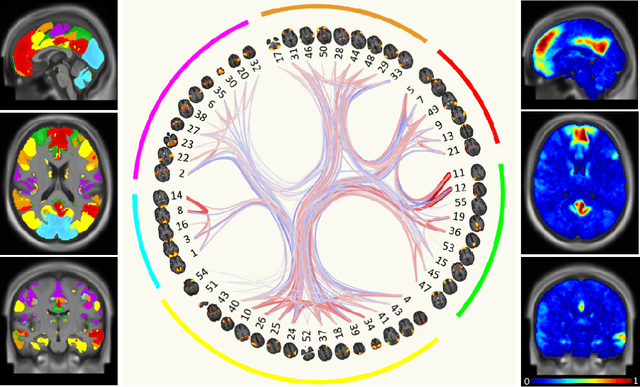
Graph convolutional networks (GCNs) allow to apply traditional convolution operations in non-Euclidean domains, where data are commonly modelled as irregular graphs. Medical imaging and, in particular, neuroscience studies often rely on such graph representations, with brain connectivity networks being a characteristic example, while ultimately seeking the locus of phenotypic or disease-related differences in the brain. These regions of interest (ROIs) are, then, considered to be closely associated with function and/or behaviour. Driven by this, we explore GCNs for the task of ROI identification and propose a visual attribution method based on class activation mapping. By undertaking a sex classification task as proof of concept, we show that this method can be used to identify salient nodes (brain regions) without prior node labels. Based on experiments conducted on neuroimaging data of more than 5000 participants from UK Biobank, we demonstrate the robustness of the proposed method in highlighting reproducible regions across individuals. We further evaluate the neurobiological relevance of the identified regions based on evidence from large-scale UK Biobank studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge